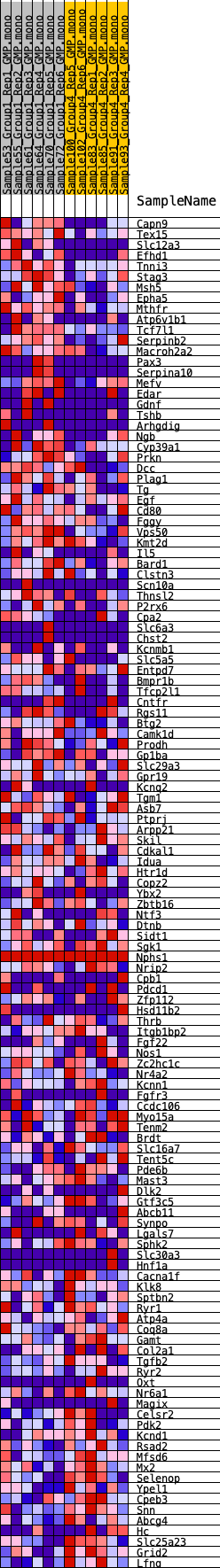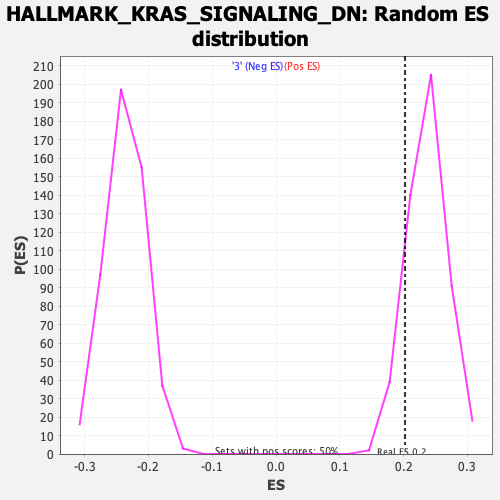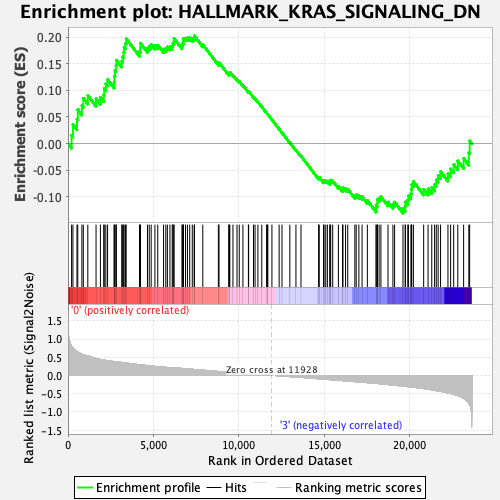
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group4.GMP.mono_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.20227905 |
| Normalized Enrichment Score (NES) | 0.85903376 |
| Nominal p-value | 0.8545455 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Capn9 | 209 | 0.802 | 0.0159 | Yes |
| 2 | Tex15 | 289 | 0.753 | 0.0357 | Yes |
| 3 | Slc12a3 | 526 | 0.653 | 0.0459 | Yes |
| 4 | Efhd1 | 571 | 0.637 | 0.0637 | Yes |
| 5 | Tnni3 | 812 | 0.580 | 0.0714 | Yes |
| 6 | Stag3 | 896 | 0.566 | 0.0853 | Yes |
| 7 | Msh5 | 1159 | 0.531 | 0.0906 | Yes |
| 8 | Epha5 | 1641 | 0.463 | 0.0844 | Yes |
| 9 | Mthfr | 1896 | 0.436 | 0.0871 | Yes |
| 10 | Atp6v1b1 | 2094 | 0.421 | 0.0917 | Yes |
| 11 | Tcf7l1 | 2117 | 0.419 | 0.1037 | Yes |
| 12 | Serpinb2 | 2213 | 0.411 | 0.1123 | Yes |
| 13 | Macroh2a2 | 2315 | 0.404 | 0.1205 | Yes |
| 14 | Pax3 | 2699 | 0.379 | 0.1159 | Yes |
| 15 | Serpina10 | 2708 | 0.379 | 0.1273 | Yes |
| 16 | Mefv | 2749 | 0.378 | 0.1372 | Yes |
| 17 | Edar | 2806 | 0.372 | 0.1463 | Yes |
| 18 | Gdnf | 2831 | 0.370 | 0.1567 | Yes |
| 19 | Tshb | 3142 | 0.354 | 0.1545 | Yes |
| 20 | Arhgdig | 3197 | 0.352 | 0.1630 | Yes |
| 21 | Ngb | 3257 | 0.349 | 0.1713 | Yes |
| 22 | Cyp39a1 | 3280 | 0.347 | 0.1811 | Yes |
| 23 | Prkn | 3366 | 0.342 | 0.1880 | Yes |
| 24 | Dcc | 3401 | 0.340 | 0.1971 | Yes |
| 25 | Plag1 | 4183 | 0.293 | 0.1729 | Yes |
| 26 | Tg | 4234 | 0.291 | 0.1798 | Yes |
| 27 | Egf | 4237 | 0.291 | 0.1887 | Yes |
| 28 | Cd80 | 4649 | 0.268 | 0.1794 | Yes |
| 29 | Fggy | 4752 | 0.263 | 0.1832 | Yes |
| 30 | Vps50 | 4866 | 0.257 | 0.1863 | Yes |
| 31 | Kmt2d | 5085 | 0.247 | 0.1847 | Yes |
| 32 | Il5 | 5251 | 0.238 | 0.1850 | Yes |
| 33 | Bard1 | 5591 | 0.228 | 0.1776 | Yes |
| 34 | Clstn3 | 5722 | 0.221 | 0.1789 | Yes |
| 35 | Scn10a | 5813 | 0.218 | 0.1818 | Yes |
| 36 | Thnsl2 | 5963 | 0.214 | 0.1821 | Yes |
| 37 | P2rx6 | 6097 | 0.209 | 0.1829 | Yes |
| 38 | Cpa2 | 6137 | 0.206 | 0.1876 | Yes |
| 39 | Slc6a3 | 6194 | 0.204 | 0.1915 | Yes |
| 40 | Chst2 | 6199 | 0.204 | 0.1976 | Yes |
| 41 | Kcnmb1 | 6668 | 0.192 | 0.1836 | Yes |
| 42 | Slc5a5 | 6703 | 0.190 | 0.1881 | Yes |
| 43 | Entpd7 | 6750 | 0.188 | 0.1919 | Yes |
| 44 | Bmpr1b | 6755 | 0.188 | 0.1976 | Yes |
| 45 | Tfcp2l1 | 6875 | 0.182 | 0.1981 | Yes |
| 46 | Cntfr | 6987 | 0.178 | 0.1989 | Yes |
| 47 | Rgs11 | 7120 | 0.174 | 0.1986 | Yes |
| 48 | Btg2 | 7271 | 0.168 | 0.1974 | Yes |
| 49 | Camk1d | 7383 | 0.164 | 0.1978 | Yes |
| 50 | Prodh | 7396 | 0.163 | 0.2023 | Yes |
| 51 | Gp1ba | 7886 | 0.145 | 0.1860 | No |
| 52 | Slc29a3 | 8796 | 0.108 | 0.1506 | No |
| 53 | Gpr19 | 8835 | 0.107 | 0.1523 | No |
| 54 | Kcnq2 | 9399 | 0.088 | 0.1311 | No |
| 55 | Tgm1 | 9455 | 0.086 | 0.1314 | No |
| 56 | Asb7 | 9461 | 0.086 | 0.1338 | No |
| 57 | Ptprj | 9650 | 0.079 | 0.1283 | No |
| 58 | Arpp21 | 9887 | 0.071 | 0.1204 | No |
| 59 | Skil | 10018 | 0.065 | 0.1169 | No |
| 60 | Cdkal1 | 10233 | 0.058 | 0.1096 | No |
| 61 | Idua | 10554 | 0.046 | 0.0974 | No |
| 62 | Htr1d | 10568 | 0.045 | 0.0983 | No |
| 63 | Copz2 | 10852 | 0.035 | 0.0873 | No |
| 64 | Ybx2 | 10943 | 0.031 | 0.0844 | No |
| 65 | Zbtb16 | 11110 | 0.026 | 0.0782 | No |
| 66 | Ntf3 | 11336 | 0.020 | 0.0692 | No |
| 67 | Dtnb | 11617 | 0.011 | 0.0576 | No |
| 68 | Sidt1 | 11666 | 0.009 | 0.0558 | No |
| 69 | Sgk1 | 11692 | 0.008 | 0.0550 | No |
| 70 | Nphs1 | 11930 | 0.000 | 0.0449 | No |
| 71 | Nrip2 | 12352 | -0.006 | 0.0272 | No |
| 72 | Cpb1 | 12519 | -0.012 | 0.0205 | No |
| 73 | Pdcd1 | 12978 | -0.028 | 0.0019 | No |
| 74 | Zfp112 | 13335 | -0.040 | -0.0121 | No |
| 75 | Hsd11b2 | 13630 | -0.051 | -0.0230 | No |
| 76 | Thrb | 14661 | -0.086 | -0.0642 | No |
| 77 | Itgb1bp2 | 14698 | -0.088 | -0.0630 | No |
| 78 | Fgf22 | 14947 | -0.096 | -0.0706 | No |
| 79 | Nos1 | 14981 | -0.098 | -0.0690 | No |
| 80 | Zc2hc1c | 15079 | -0.101 | -0.0699 | No |
| 81 | Nr4a2 | 15174 | -0.105 | -0.0707 | No |
| 82 | Kcnn1 | 15321 | -0.111 | -0.0735 | No |
| 83 | Fgfr3 | 15327 | -0.111 | -0.0703 | No |
| 84 | Ccdc106 | 15368 | -0.112 | -0.0685 | No |
| 85 | Myo15a | 15486 | -0.116 | -0.0699 | No |
| 86 | Tenm2 | 15819 | -0.129 | -0.0800 | No |
| 87 | Brdt | 16061 | -0.139 | -0.0860 | No |
| 88 | Slc16a7 | 16086 | -0.140 | -0.0827 | No |
| 89 | Tent5c | 16239 | -0.145 | -0.0847 | No |
| 90 | Pde6b | 16366 | -0.151 | -0.0854 | No |
| 91 | Mast3 | 16782 | -0.166 | -0.0979 | No |
| 92 | Dlk2 | 16860 | -0.169 | -0.0960 | No |
| 93 | Gtf3c5 | 17017 | -0.175 | -0.0972 | No |
| 94 | Abcb11 | 17204 | -0.181 | -0.0995 | No |
| 95 | Synpo | 17513 | -0.194 | -0.1067 | No |
| 96 | Lgals7 | 18027 | -0.217 | -0.1218 | No |
| 97 | Sphk2 | 18029 | -0.217 | -0.1151 | No |
| 98 | Slc30a3 | 18102 | -0.219 | -0.1114 | No |
| 99 | Hnf1a | 18103 | -0.219 | -0.1047 | No |
| 100 | Cacna1f | 18201 | -0.220 | -0.1020 | No |
| 101 | Klk8 | 18307 | -0.225 | -0.0995 | No |
| 102 | Sptbn2 | 18721 | -0.244 | -0.1096 | No |
| 103 | Ryr1 | 19008 | -0.261 | -0.1137 | No |
| 104 | Atp4a | 19107 | -0.266 | -0.1096 | No |
| 105 | Coq8a | 19597 | -0.287 | -0.1216 | No |
| 106 | Gamt | 19727 | -0.296 | -0.1179 | No |
| 107 | Col2a1 | 19737 | -0.296 | -0.1092 | No |
| 108 | Tgfb2 | 19879 | -0.305 | -0.1057 | No |
| 109 | Ryr2 | 19915 | -0.307 | -0.0978 | No |
| 110 | Oxt | 20062 | -0.313 | -0.0943 | No |
| 111 | Nr6a1 | 20094 | -0.314 | -0.0860 | No |
| 112 | Magix | 20109 | -0.314 | -0.0768 | No |
| 113 | Celsr2 | 20207 | -0.320 | -0.0711 | No |
| 114 | Pdk2 | 20807 | -0.357 | -0.0856 | No |
| 115 | Kcnd1 | 21065 | -0.374 | -0.0849 | No |
| 116 | Rsad2 | 21281 | -0.391 | -0.0820 | No |
| 117 | Mfsd6 | 21447 | -0.405 | -0.0765 | No |
| 118 | Mx2 | 21545 | -0.413 | -0.0679 | No |
| 119 | Selenop | 21654 | -0.421 | -0.0595 | No |
| 120 | Ypel1 | 21795 | -0.430 | -0.0522 | No |
| 121 | Cpeb3 | 22228 | -0.475 | -0.0559 | No |
| 122 | Snn | 22386 | -0.492 | -0.0474 | No |
| 123 | Abcg4 | 22564 | -0.513 | -0.0391 | No |
| 124 | Hc | 22804 | -0.547 | -0.0323 | No |
| 125 | Slc25a23 | 23145 | -0.622 | -0.0276 | No |
| 126 | Grid2 | 23450 | -0.757 | -0.0172 | No |
| 127 | Lfng | 23498 | -0.794 | 0.0054 | No |