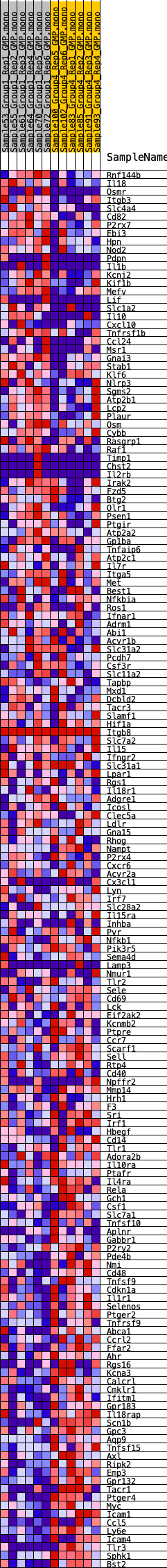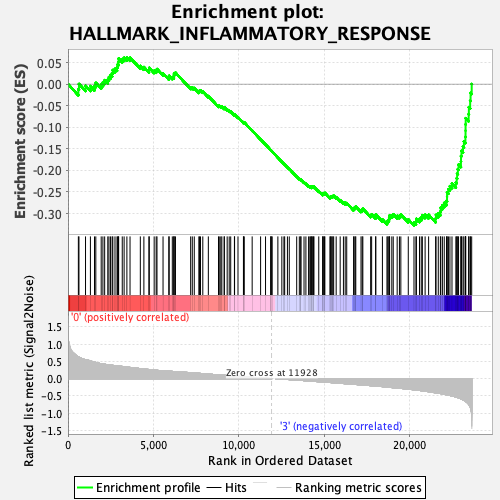
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group4.GMP.mono_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | -0.3278704 |
| Normalized Enrichment Score (NES) | -1.4495631 |
| Nominal p-value | 0.022088353 |
| FDR q-value | 0.23665108 |
| FWER p-Value | 0.496 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rnf144b | 606 | 0.628 | -0.0119 | No |
| 2 | Il18 | 641 | 0.619 | 0.0004 | No |
| 3 | Osmr | 1025 | 0.548 | -0.0038 | No |
| 4 | Itgb3 | 1311 | 0.514 | -0.0046 | No |
| 5 | Slc4a4 | 1551 | 0.476 | -0.0042 | No |
| 6 | Cd82 | 1627 | 0.466 | 0.0029 | No |
| 7 | P2rx7 | 1942 | 0.433 | -0.0009 | No |
| 8 | Ebi3 | 2042 | 0.425 | 0.0043 | No |
| 9 | Hpn | 2145 | 0.416 | 0.0092 | No |
| 10 | Nod2 | 2328 | 0.403 | 0.0104 | No |
| 11 | Pdpn | 2396 | 0.399 | 0.0164 | No |
| 12 | Il1b | 2489 | 0.391 | 0.0211 | No |
| 13 | Kcnj2 | 2582 | 0.387 | 0.0258 | No |
| 14 | Kif1b | 2621 | 0.384 | 0.0327 | No |
| 15 | Mefv | 2749 | 0.378 | 0.0356 | No |
| 16 | Lif | 2868 | 0.370 | 0.0388 | No |
| 17 | Slc1a2 | 2902 | 0.370 | 0.0456 | No |
| 18 | Il10 | 2955 | 0.368 | 0.0515 | No |
| 19 | Cxcl10 | 2962 | 0.367 | 0.0594 | No |
| 20 | Tnfrsf1b | 3174 | 0.352 | 0.0582 | No |
| 21 | Ccl24 | 3283 | 0.347 | 0.0613 | No |
| 22 | Msr1 | 3445 | 0.337 | 0.0619 | No |
| 23 | Gnai3 | 3629 | 0.324 | 0.0613 | No |
| 24 | Stab1 | 4231 | 0.291 | 0.0421 | No |
| 25 | Klf6 | 4439 | 0.279 | 0.0395 | No |
| 26 | Nlrp3 | 4734 | 0.264 | 0.0328 | No |
| 27 | Sgms2 | 4753 | 0.263 | 0.0379 | No |
| 28 | Atp2b1 | 5041 | 0.249 | 0.0312 | No |
| 29 | Lcp2 | 5157 | 0.243 | 0.0316 | No |
| 30 | Plaur | 5206 | 0.240 | 0.0349 | No |
| 31 | Osm | 5560 | 0.230 | 0.0250 | No |
| 32 | Cybb | 5899 | 0.217 | 0.0154 | No |
| 33 | Rasgrp1 | 5917 | 0.216 | 0.0194 | No |
| 34 | Raf1 | 6102 | 0.208 | 0.0162 | No |
| 35 | Timp1 | 6191 | 0.204 | 0.0170 | No |
| 36 | Chst2 | 6199 | 0.204 | 0.0212 | No |
| 37 | Il2rb | 6208 | 0.204 | 0.0254 | No |
| 38 | Irak2 | 6287 | 0.202 | 0.0266 | No |
| 39 | Fzd5 | 7176 | 0.172 | -0.0075 | No |
| 40 | Btg2 | 7271 | 0.168 | -0.0078 | No |
| 41 | Olr1 | 7387 | 0.163 | -0.0091 | No |
| 42 | Psen1 | 7657 | 0.154 | -0.0171 | No |
| 43 | Ptgir | 7690 | 0.152 | -0.0151 | No |
| 44 | Atp2a2 | 7751 | 0.150 | -0.0143 | No |
| 45 | Gp1ba | 7886 | 0.145 | -0.0168 | No |
| 46 | Tnfaip6 | 8210 | 0.131 | -0.0277 | No |
| 47 | Atp2c1 | 8804 | 0.108 | -0.0506 | No |
| 48 | Il7r | 8848 | 0.106 | -0.0500 | No |
| 49 | Itga5 | 8938 | 0.104 | -0.0515 | No |
| 50 | Met | 8999 | 0.102 | -0.0518 | No |
| 51 | Best1 | 9139 | 0.097 | -0.0556 | No |
| 52 | Nfkbia | 9143 | 0.096 | -0.0536 | No |
| 53 | Ros1 | 9312 | 0.091 | -0.0588 | No |
| 54 | Ifnar1 | 9431 | 0.087 | -0.0619 | No |
| 55 | Adrm1 | 9519 | 0.084 | -0.0637 | No |
| 56 | Abi1 | 9736 | 0.076 | -0.0712 | No |
| 57 | Acvr1b | 9745 | 0.076 | -0.0699 | No |
| 58 | Slc31a2 | 9945 | 0.069 | -0.0769 | No |
| 59 | Pcdh7 | 10263 | 0.057 | -0.0891 | No |
| 60 | Csf3r | 10269 | 0.056 | -0.0881 | No |
| 61 | Slc11a2 | 10312 | 0.055 | -0.0887 | No |
| 62 | Tapbp | 10774 | 0.037 | -0.1075 | No |
| 63 | Mxd1 | 11268 | 0.022 | -0.1280 | No |
| 64 | Dcbld2 | 11556 | 0.013 | -0.1400 | No |
| 65 | Tacr3 | 11861 | 0.002 | -0.1529 | No |
| 66 | Slamf1 | 11871 | 0.002 | -0.1532 | No |
| 67 | Hif1a | 11894 | 0.001 | -0.1541 | No |
| 68 | Itgb8 | 11945 | 0.000 | -0.1562 | No |
| 69 | Slc7a2 | 12278 | -0.003 | -0.1703 | No |
| 70 | Il15 | 12510 | -0.011 | -0.1799 | No |
| 71 | Ifngr2 | 12632 | -0.015 | -0.1847 | No |
| 72 | Slc31a1 | 12676 | -0.017 | -0.1862 | No |
| 73 | Lpar1 | 12837 | -0.022 | -0.1925 | No |
| 74 | Rgs1 | 12945 | -0.027 | -0.1965 | No |
| 75 | Il18r1 | 13379 | -0.041 | -0.2140 | No |
| 76 | Adgre1 | 13533 | -0.047 | -0.2195 | No |
| 77 | Icosl | 13622 | -0.051 | -0.2221 | No |
| 78 | Clec5a | 13623 | -0.051 | -0.2210 | No |
| 79 | Ldlr | 13808 | -0.056 | -0.2276 | No |
| 80 | Gna15 | 13918 | -0.060 | -0.2309 | No |
| 81 | Rhog | 14074 | -0.065 | -0.2361 | No |
| 82 | Nampt | 14150 | -0.068 | -0.2378 | No |
| 83 | P2rx4 | 14179 | -0.070 | -0.2374 | No |
| 84 | Cxcr6 | 14257 | -0.072 | -0.2391 | No |
| 85 | Acvr2a | 14263 | -0.073 | -0.2377 | No |
| 86 | Cx3cl1 | 14291 | -0.074 | -0.2372 | No |
| 87 | Lyn | 14365 | -0.077 | -0.2386 | No |
| 88 | Irf7 | 14388 | -0.078 | -0.2378 | No |
| 89 | Slc28a2 | 14670 | -0.087 | -0.2479 | No |
| 90 | Il15ra | 14899 | -0.095 | -0.2555 | No |
| 91 | Inhba | 14913 | -0.095 | -0.2539 | No |
| 92 | Pvr | 14934 | -0.096 | -0.2527 | No |
| 93 | Nfkb1 | 15003 | -0.099 | -0.2534 | No |
| 94 | Pik3r5 | 15027 | -0.100 | -0.2521 | No |
| 95 | Sema4d | 15326 | -0.111 | -0.2624 | No |
| 96 | Lamp3 | 15351 | -0.112 | -0.2609 | No |
| 97 | Nmur1 | 15400 | -0.114 | -0.2605 | No |
| 98 | Tlr2 | 15447 | -0.115 | -0.2599 | No |
| 99 | Sele | 15496 | -0.116 | -0.2593 | No |
| 100 | Cd69 | 15542 | -0.118 | -0.2586 | No |
| 101 | Lck | 15689 | -0.124 | -0.2621 | No |
| 102 | Eif2ak2 | 15923 | -0.134 | -0.2691 | No |
| 103 | Kcnmb2 | 16113 | -0.140 | -0.2740 | No |
| 104 | Ptpre | 16216 | -0.145 | -0.2752 | No |
| 105 | Ccr7 | 16306 | -0.149 | -0.2757 | No |
| 106 | Scarf1 | 16707 | -0.163 | -0.2891 | No |
| 107 | Sell | 16738 | -0.164 | -0.2867 | No |
| 108 | Rtp4 | 16801 | -0.166 | -0.2857 | No |
| 109 | Cd40 | 16836 | -0.168 | -0.2834 | No |
| 110 | Npffr2 | 17142 | -0.179 | -0.2924 | No |
| 111 | Mmp14 | 17211 | -0.181 | -0.2913 | No |
| 112 | Hrh1 | 17249 | -0.183 | -0.2888 | No |
| 113 | F3 | 17706 | -0.202 | -0.3038 | No |
| 114 | Sri | 17764 | -0.205 | -0.3017 | No |
| 115 | Irf1 | 18000 | -0.215 | -0.3069 | No |
| 116 | Hbegf | 18004 | -0.215 | -0.3023 | No |
| 117 | Cd14 | 18394 | -0.230 | -0.3138 | No |
| 118 | Tlr1 | 18660 | -0.241 | -0.3197 | No |
| 119 | Adora2b | 18709 | -0.244 | -0.3164 | No |
| 120 | Il10ra | 18772 | -0.248 | -0.3135 | No |
| 121 | Ptafr | 18793 | -0.249 | -0.3088 | No |
| 122 | Il4ra | 18815 | -0.250 | -0.3042 | No |
| 123 | Rela | 18944 | -0.257 | -0.3039 | No |
| 124 | Gch1 | 19027 | -0.262 | -0.3016 | No |
| 125 | Csf1 | 19257 | -0.272 | -0.3054 | No |
| 126 | Slc7a1 | 19390 | -0.279 | -0.3048 | No |
| 127 | Tnfsf10 | 19472 | -0.284 | -0.3020 | No |
| 128 | Aplnr | 19907 | -0.306 | -0.3137 | No |
| 129 | Gabbr1 | 20241 | -0.322 | -0.3207 | Yes |
| 130 | P2ry2 | 20357 | -0.328 | -0.3184 | Yes |
| 131 | Pde4b | 20381 | -0.330 | -0.3120 | Yes |
| 132 | Nmi | 20565 | -0.341 | -0.3123 | Yes |
| 133 | Cd48 | 20665 | -0.347 | -0.3088 | Yes |
| 134 | Tnfsf9 | 20731 | -0.352 | -0.3038 | Yes |
| 135 | Cdkn1a | 20892 | -0.363 | -0.3026 | Yes |
| 136 | Il1r1 | 21097 | -0.377 | -0.3029 | Yes |
| 137 | Selenos | 21508 | -0.410 | -0.3113 | Yes |
| 138 | Ptger2 | 21528 | -0.412 | -0.3030 | Yes |
| 139 | Tnfrsf9 | 21662 | -0.421 | -0.2993 | Yes |
| 140 | Abca1 | 21785 | -0.429 | -0.2950 | Yes |
| 141 | Ccrl2 | 21807 | -0.432 | -0.2863 | Yes |
| 142 | Ffar2 | 21903 | -0.441 | -0.2806 | Yes |
| 143 | Ahr | 22011 | -0.454 | -0.2751 | Yes |
| 144 | Rgs16 | 22145 | -0.467 | -0.2704 | Yes |
| 145 | Kcna3 | 22175 | -0.470 | -0.2612 | Yes |
| 146 | Calcrl | 22177 | -0.471 | -0.2508 | Yes |
| 147 | Cmklr1 | 22256 | -0.480 | -0.2435 | Yes |
| 148 | Ifitm1 | 22344 | -0.488 | -0.2364 | Yes |
| 149 | Gpr183 | 22464 | -0.500 | -0.2304 | Yes |
| 150 | Il18rap | 22691 | -0.530 | -0.2283 | Yes |
| 151 | Scn1b | 22738 | -0.537 | -0.2183 | Yes |
| 152 | Gpc3 | 22765 | -0.540 | -0.2075 | Yes |
| 153 | Aqp9 | 22807 | -0.548 | -0.1970 | Yes |
| 154 | Tnfsf15 | 22848 | -0.554 | -0.1865 | Yes |
| 155 | Axl | 22987 | -0.585 | -0.1794 | Yes |
| 156 | Ripk2 | 22993 | -0.586 | -0.1666 | Yes |
| 157 | Emp3 | 23019 | -0.592 | -0.1545 | Yes |
| 158 | Gpr132 | 23109 | -0.616 | -0.1447 | Yes |
| 159 | Tacr1 | 23162 | -0.628 | -0.1330 | Yes |
| 160 | Ptger4 | 23249 | -0.654 | -0.1221 | Yes |
| 161 | Myc | 23257 | -0.657 | -0.1079 | Yes |
| 162 | Icam1 | 23259 | -0.657 | -0.0934 | Yes |
| 163 | Ccl5 | 23266 | -0.660 | -0.0790 | Yes |
| 164 | Ly6e | 23439 | -0.750 | -0.0697 | Yes |
| 165 | Icam4 | 23452 | -0.759 | -0.0534 | Yes |
| 166 | Tlr3 | 23528 | -0.819 | -0.0384 | Yes |
| 167 | Sphk1 | 23552 | -0.854 | -0.0204 | Yes |
| 168 | Bst2 | 23617 | -1.059 | 0.0003 | Yes |