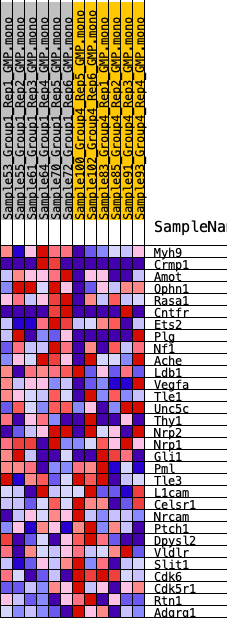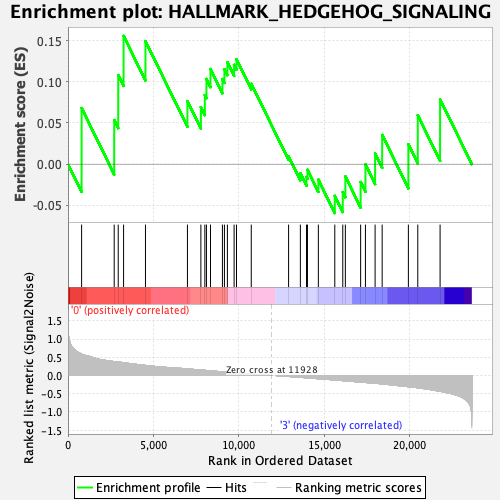
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group4.GMP.mono_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.15573552 |
| Normalized Enrichment Score (NES) | 0.5232966 |
| Nominal p-value | 0.9791271 |
| FDR q-value | 0.9719003 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Myh9 | 796 | 0.584 | 0.0682 | Yes |
| 2 | Crmp1 | 2703 | 0.379 | 0.0535 | Yes |
| 3 | Amot | 2938 | 0.369 | 0.1081 | Yes |
| 4 | Ophn1 | 3252 | 0.349 | 0.1557 | Yes |
| 5 | Rasa1 | 4532 | 0.274 | 0.1494 | No |
| 6 | Cntfr | 6987 | 0.178 | 0.0765 | No |
| 7 | Ets2 | 7774 | 0.149 | 0.0692 | No |
| 8 | Plg | 8006 | 0.140 | 0.0838 | No |
| 9 | Nf1 | 8099 | 0.135 | 0.1035 | No |
| 10 | Ache | 8336 | 0.126 | 0.1155 | No |
| 11 | Ldb1 | 9034 | 0.100 | 0.1034 | No |
| 12 | Vegfa | 9151 | 0.096 | 0.1153 | No |
| 13 | Tle1 | 9325 | 0.090 | 0.1237 | No |
| 14 | Unc5c | 9718 | 0.077 | 0.1206 | No |
| 15 | Thy1 | 9857 | 0.072 | 0.1273 | No |
| 16 | Nrp2 | 10721 | 0.039 | 0.0977 | No |
| 17 | Nrp1 | 12905 | -0.025 | 0.0095 | No |
| 18 | Gli1 | 13594 | -0.050 | -0.0110 | No |
| 19 | Pml | 13954 | -0.061 | -0.0155 | No |
| 20 | Tle3 | 14009 | -0.063 | -0.0068 | No |
| 21 | L1cam | 14646 | -0.086 | -0.0187 | No |
| 22 | Celsr1 | 15608 | -0.120 | -0.0384 | No |
| 23 | Nrcam | 16075 | -0.139 | -0.0339 | No |
| 24 | Ptch1 | 16226 | -0.145 | -0.0149 | No |
| 25 | Dpysl2 | 17120 | -0.178 | -0.0217 | No |
| 26 | Vldlr | 17401 | -0.190 | -0.0004 | No |
| 27 | Slit1 | 17966 | -0.214 | 0.0130 | No |
| 28 | Cdk6 | 18379 | -0.229 | 0.0355 | No |
| 29 | Cdk5r1 | 19913 | -0.307 | 0.0241 | No |
| 30 | Rtn1 | 20463 | -0.335 | 0.0593 | No |
| 31 | Adgrg1 | 21768 | -0.427 | 0.0787 | No |