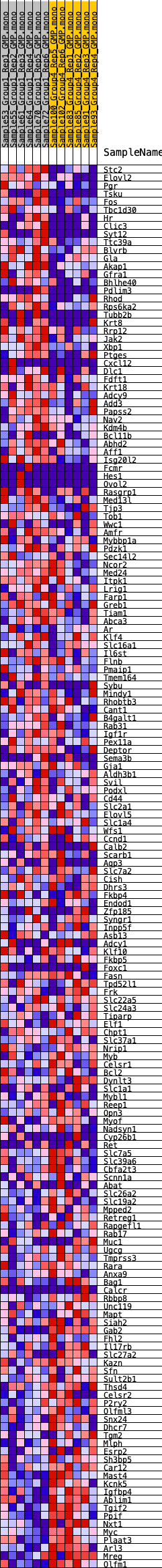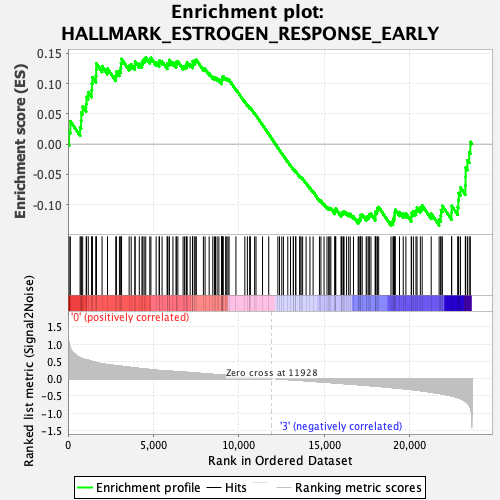
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group4.GMP.mono_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_ESTROGEN_RESPONSE_EARLY |
| Enrichment Score (ES) | 0.14315124 |
| Normalized Enrichment Score (NES) | 0.6470558 |
| Nominal p-value | 0.996063 |
| FDR q-value | 0.9842386 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Stc2 | 66 | 1.008 | 0.0207 | Yes |
| 2 | Elovl2 | 137 | 0.869 | 0.0379 | Yes |
| 3 | Pgr | 708 | 0.604 | 0.0277 | Yes |
| 4 | Tsku | 765 | 0.591 | 0.0391 | Yes |
| 5 | Fos | 777 | 0.588 | 0.0523 | Yes |
| 6 | Tbc1d30 | 858 | 0.570 | 0.0622 | Yes |
| 7 | Hr | 1064 | 0.544 | 0.0661 | Yes |
| 8 | Clic3 | 1082 | 0.541 | 0.0780 | Yes |
| 9 | Syt12 | 1185 | 0.529 | 0.0860 | Yes |
| 10 | Ttc39a | 1385 | 0.500 | 0.0891 | Yes |
| 11 | Blvrb | 1402 | 0.497 | 0.1000 | Yes |
| 12 | Gla | 1428 | 0.495 | 0.1105 | Yes |
| 13 | Akap1 | 1635 | 0.464 | 0.1125 | Yes |
| 14 | Gfra1 | 1650 | 0.463 | 0.1227 | Yes |
| 15 | Bhlhe40 | 1657 | 0.462 | 0.1332 | Yes |
| 16 | Pdlim3 | 1993 | 0.427 | 0.1289 | Yes |
| 17 | Rhod | 2312 | 0.404 | 0.1248 | Yes |
| 18 | Rps6ka2 | 2795 | 0.373 | 0.1129 | Yes |
| 19 | Tubb2b | 2833 | 0.370 | 0.1200 | Yes |
| 20 | Krt8 | 3008 | 0.362 | 0.1210 | Yes |
| 21 | Rrp12 | 3089 | 0.359 | 0.1259 | Yes |
| 22 | Jak2 | 3102 | 0.358 | 0.1338 | Yes |
| 23 | Xbp1 | 3134 | 0.355 | 0.1407 | Yes |
| 24 | Ptges | 3577 | 0.328 | 0.1295 | Yes |
| 25 | Cxcl12 | 3697 | 0.320 | 0.1319 | Yes |
| 26 | Dlc1 | 3914 | 0.308 | 0.1298 | Yes |
| 27 | Fdft1 | 3924 | 0.307 | 0.1366 | Yes |
| 28 | Krt18 | 4171 | 0.294 | 0.1330 | Yes |
| 29 | Adcy9 | 4327 | 0.285 | 0.1330 | Yes |
| 30 | Add3 | 4376 | 0.282 | 0.1375 | Yes |
| 31 | Papss2 | 4456 | 0.279 | 0.1407 | Yes |
| 32 | Nav2 | 4548 | 0.273 | 0.1432 | Yes |
| 33 | Kdm4b | 4785 | 0.261 | 0.1392 | No |
| 34 | Bcl11b | 4850 | 0.258 | 0.1425 | No |
| 35 | Abhd2 | 5156 | 0.243 | 0.1351 | No |
| 36 | Aff1 | 5331 | 0.234 | 0.1331 | No |
| 37 | Isg20l2 | 5347 | 0.233 | 0.1379 | No |
| 38 | Fcmr | 5503 | 0.230 | 0.1367 | No |
| 39 | Hes1 | 5802 | 0.218 | 0.1290 | No |
| 40 | Ovol2 | 5815 | 0.218 | 0.1336 | No |
| 41 | Rasgrp1 | 5917 | 0.216 | 0.1343 | No |
| 42 | Med13l | 5925 | 0.216 | 0.1391 | No |
| 43 | Tjp3 | 6141 | 0.206 | 0.1347 | No |
| 44 | Tob1 | 6329 | 0.201 | 0.1314 | No |
| 45 | Wwc1 | 6332 | 0.201 | 0.1360 | No |
| 46 | Amfr | 6427 | 0.197 | 0.1366 | No |
| 47 | Mybbp1a | 6733 | 0.189 | 0.1280 | No |
| 48 | Pdzk1 | 6820 | 0.185 | 0.1286 | No |
| 49 | Sec14l2 | 6913 | 0.181 | 0.1289 | No |
| 50 | Ncor2 | 6941 | 0.180 | 0.1320 | No |
| 51 | Med24 | 6967 | 0.179 | 0.1351 | No |
| 52 | Itpk1 | 7146 | 0.172 | 0.1315 | No |
| 53 | Lrig1 | 7285 | 0.167 | 0.1295 | No |
| 54 | Farp1 | 7292 | 0.167 | 0.1331 | No |
| 55 | Greb1 | 7293 | 0.167 | 0.1370 | No |
| 56 | Tiam1 | 7426 | 0.161 | 0.1351 | No |
| 57 | Abca3 | 7429 | 0.161 | 0.1388 | No |
| 58 | Ar | 7507 | 0.159 | 0.1392 | No |
| 59 | Klf4 | 7921 | 0.143 | 0.1250 | No |
| 60 | Slc16a1 | 8019 | 0.139 | 0.1241 | No |
| 61 | Il6st | 8258 | 0.129 | 0.1169 | No |
| 62 | Flnb | 8471 | 0.121 | 0.1107 | No |
| 63 | Pmaip1 | 8566 | 0.117 | 0.1094 | No |
| 64 | Tmem164 | 8628 | 0.115 | 0.1095 | No |
| 65 | Sybu | 8724 | 0.110 | 0.1080 | No |
| 66 | Mindy1 | 8827 | 0.107 | 0.1062 | No |
| 67 | Rhobtb3 | 8992 | 0.102 | 0.1015 | No |
| 68 | Cant1 | 8993 | 0.102 | 0.1039 | No |
| 69 | B4galt1 | 8997 | 0.102 | 0.1062 | No |
| 70 | Rab31 | 9019 | 0.100 | 0.1076 | No |
| 71 | Igf1r | 9022 | 0.100 | 0.1099 | No |
| 72 | Pex11a | 9051 | 0.099 | 0.1110 | No |
| 73 | Deptor | 9081 | 0.098 | 0.1120 | No |
| 74 | Sema3b | 9222 | 0.094 | 0.1082 | No |
| 75 | Gja1 | 9287 | 0.092 | 0.1076 | No |
| 76 | Aldh3b1 | 9359 | 0.089 | 0.1067 | No |
| 77 | Svil | 9422 | 0.088 | 0.1061 | No |
| 78 | Podxl | 9825 | 0.073 | 0.0907 | No |
| 79 | Cd44 | 10345 | 0.054 | 0.0698 | No |
| 80 | Slc2a1 | 10494 | 0.048 | 0.0646 | No |
| 81 | Elovl5 | 10641 | 0.043 | 0.0594 | No |
| 82 | Slc1a4 | 10644 | 0.042 | 0.0603 | No |
| 83 | Wfs1 | 10661 | 0.042 | 0.0606 | No |
| 84 | Ccnd1 | 10919 | 0.032 | 0.0503 | No |
| 85 | Calb2 | 10999 | 0.029 | 0.0477 | No |
| 86 | Scarb1 | 11381 | 0.018 | 0.0318 | No |
| 87 | Aqp3 | 11743 | 0.006 | 0.0166 | No |
| 88 | Slc7a2 | 12278 | -0.003 | -0.0061 | No |
| 89 | Cish | 12372 | -0.006 | -0.0099 | No |
| 90 | Dhrs3 | 12506 | -0.011 | -0.0153 | No |
| 91 | Fkbp4 | 12603 | -0.014 | -0.0191 | No |
| 92 | Endod1 | 12858 | -0.023 | -0.0294 | No |
| 93 | Zfp185 | 13019 | -0.029 | -0.0355 | No |
| 94 | Syngr1 | 13183 | -0.035 | -0.0417 | No |
| 95 | Inpp5f | 13185 | -0.035 | -0.0409 | No |
| 96 | Asb13 | 13319 | -0.039 | -0.0456 | No |
| 97 | Adcy1 | 13328 | -0.040 | -0.0451 | No |
| 98 | Klf10 | 13554 | -0.048 | -0.0535 | No |
| 99 | Fkbp5 | 13573 | -0.049 | -0.0532 | No |
| 100 | Foxc1 | 13638 | -0.051 | -0.0547 | No |
| 101 | Fasn | 13726 | -0.053 | -0.0572 | No |
| 102 | Tpd52l1 | 13929 | -0.060 | -0.0644 | No |
| 103 | Frk | 14155 | -0.069 | -0.0724 | No |
| 104 | Slc22a5 | 14340 | -0.076 | -0.0785 | No |
| 105 | Slc24a3 | 14711 | -0.088 | -0.0922 | No |
| 106 | Tiparp | 14797 | -0.091 | -0.0937 | No |
| 107 | Elf1 | 14979 | -0.098 | -0.0991 | No |
| 108 | Chpt1 | 15152 | -0.105 | -0.1040 | No |
| 109 | Slc37a1 | 15249 | -0.108 | -0.1056 | No |
| 110 | Nrip1 | 15314 | -0.110 | -0.1058 | No |
| 111 | Myb | 15378 | -0.113 | -0.1058 | No |
| 112 | Celsr1 | 15608 | -0.120 | -0.1128 | No |
| 113 | Bcl2 | 15620 | -0.121 | -0.1104 | No |
| 114 | Dynlt3 | 15658 | -0.123 | -0.1092 | No |
| 115 | Slc1a1 | 15660 | -0.123 | -0.1063 | No |
| 116 | Mybl1 | 15974 | -0.136 | -0.1165 | No |
| 117 | Reep1 | 16007 | -0.137 | -0.1147 | No |
| 118 | Opn3 | 16031 | -0.138 | -0.1125 | No |
| 119 | Myof | 16117 | -0.141 | -0.1128 | No |
| 120 | Nadsyn1 | 16150 | -0.142 | -0.1109 | No |
| 121 | Cyp26b1 | 16298 | -0.148 | -0.1137 | No |
| 122 | Ret | 16421 | -0.153 | -0.1153 | No |
| 123 | Slc7a5 | 16511 | -0.157 | -0.1155 | No |
| 124 | Slc39a6 | 16700 | -0.163 | -0.1197 | No |
| 125 | Cbfa2t3 | 16974 | -0.173 | -0.1273 | No |
| 126 | Scnn1a | 17012 | -0.175 | -0.1248 | No |
| 127 | Abat | 17080 | -0.177 | -0.1235 | No |
| 128 | Slc26a2 | 17118 | -0.178 | -0.1209 | No |
| 129 | Slc19a2 | 17128 | -0.178 | -0.1172 | No |
| 130 | Mpped2 | 17212 | -0.182 | -0.1165 | No |
| 131 | Retreg1 | 17444 | -0.191 | -0.1219 | No |
| 132 | Rapgefl1 | 17490 | -0.193 | -0.1193 | No |
| 133 | Rab17 | 17594 | -0.197 | -0.1191 | No |
| 134 | Muc1 | 17628 | -0.198 | -0.1159 | No |
| 135 | Ugcg | 17723 | -0.203 | -0.1151 | No |
| 136 | Tmprss3 | 17976 | -0.214 | -0.1209 | No |
| 137 | Rara | 17978 | -0.214 | -0.1159 | No |
| 138 | Anxa9 | 17991 | -0.215 | -0.1114 | No |
| 139 | Bag1 | 18076 | -0.218 | -0.1100 | No |
| 140 | Calcr | 18098 | -0.219 | -0.1058 | No |
| 141 | Rbbp8 | 18176 | -0.219 | -0.1039 | No |
| 142 | Unc119 | 18897 | -0.255 | -0.1287 | No |
| 143 | Mapt | 19011 | -0.261 | -0.1274 | No |
| 144 | Siah2 | 19045 | -0.263 | -0.1227 | No |
| 145 | Gab2 | 19099 | -0.265 | -0.1188 | No |
| 146 | Fhl2 | 19113 | -0.266 | -0.1131 | No |
| 147 | Il17rb | 19148 | -0.268 | -0.1084 | No |
| 148 | Slc27a2 | 19394 | -0.279 | -0.1123 | No |
| 149 | Kazn | 19612 | -0.288 | -0.1148 | No |
| 150 | Sfn | 19767 | -0.298 | -0.1144 | No |
| 151 | Sult2b1 | 20085 | -0.313 | -0.1207 | No |
| 152 | Thsd4 | 20089 | -0.313 | -0.1135 | No |
| 153 | Celsr2 | 20207 | -0.320 | -0.1110 | No |
| 154 | P2ry2 | 20357 | -0.328 | -0.1097 | No |
| 155 | Olfml3 | 20419 | -0.332 | -0.1046 | No |
| 156 | Snx24 | 20613 | -0.344 | -0.1048 | No |
| 157 | Dhcr7 | 20720 | -0.351 | -0.1012 | No |
| 158 | Tgm2 | 21248 | -0.389 | -0.1146 | No |
| 159 | Mlph | 21713 | -0.425 | -0.1245 | No |
| 160 | Esrp2 | 21805 | -0.431 | -0.1183 | No |
| 161 | Sh3bp5 | 21822 | -0.434 | -0.1089 | No |
| 162 | Car12 | 21902 | -0.441 | -0.1020 | No |
| 163 | Mast4 | 22438 | -0.496 | -0.1132 | No |
| 164 | Kcnk5 | 22448 | -0.497 | -0.1020 | No |
| 165 | Igfbp4 | 22799 | -0.547 | -0.1042 | No |
| 166 | Ablim1 | 22834 | -0.552 | -0.0928 | No |
| 167 | Tgif2 | 22857 | -0.555 | -0.0808 | No |
| 168 | Ppif | 22960 | -0.578 | -0.0716 | No |
| 169 | Nxt1 | 23245 | -0.654 | -0.0685 | No |
| 170 | Myc | 23257 | -0.657 | -0.0537 | No |
| 171 | Plaat3 | 23263 | -0.659 | -0.0386 | No |
| 172 | Arl3 | 23368 | -0.700 | -0.0267 | No |
| 173 | Mreg | 23472 | -0.771 | -0.0131 | No |
| 174 | Olfm1 | 23539 | -0.839 | 0.0036 | No |