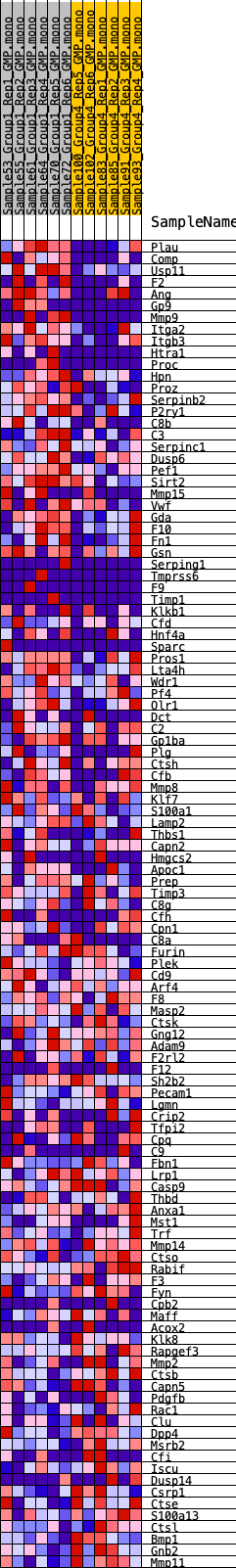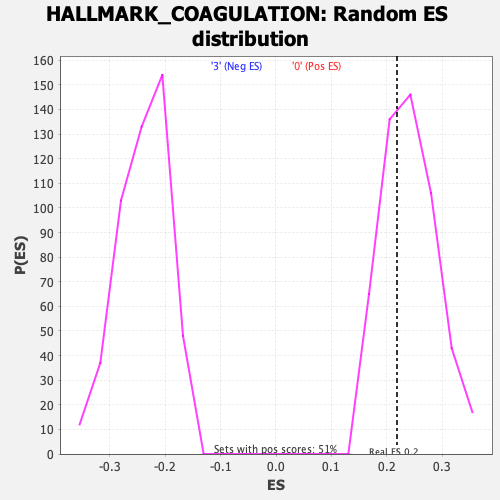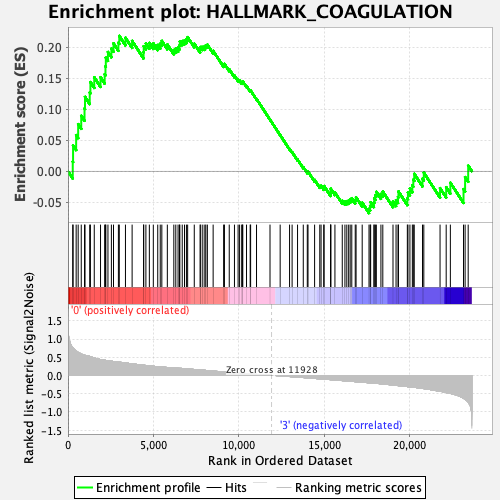
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group4.GMP.mono_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.21839982 |
| Normalized Enrichment Score (NES) | 0.9064251 |
| Nominal p-value | 0.64912283 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Plau | 278 | 0.760 | 0.0155 | Yes |
| 2 | Comp | 297 | 0.749 | 0.0417 | Yes |
| 3 | Usp11 | 477 | 0.673 | 0.0583 | Yes |
| 4 | F2 | 595 | 0.630 | 0.0760 | Yes |
| 5 | Ang | 775 | 0.589 | 0.0895 | Yes |
| 6 | Gp9 | 967 | 0.554 | 0.1013 | Yes |
| 7 | Mmp9 | 995 | 0.549 | 0.1200 | Yes |
| 8 | Itga2 | 1271 | 0.521 | 0.1270 | Yes |
| 9 | Itgb3 | 1311 | 0.514 | 0.1438 | Yes |
| 10 | Htra1 | 1536 | 0.479 | 0.1515 | Yes |
| 11 | Proc | 1903 | 0.436 | 0.1517 | Yes |
| 12 | Hpn | 2145 | 0.416 | 0.1564 | Yes |
| 13 | Proz | 2192 | 0.413 | 0.1692 | Yes |
| 14 | Serpinb2 | 2213 | 0.411 | 0.1832 | Yes |
| 15 | P2ry1 | 2337 | 0.402 | 0.1924 | Yes |
| 16 | C8b | 2538 | 0.390 | 0.1979 | Yes |
| 17 | C3 | 2662 | 0.380 | 0.2064 | Yes |
| 18 | Serpinc1 | 2948 | 0.368 | 0.2075 | Yes |
| 19 | Dusp6 | 3000 | 0.363 | 0.2184 | Yes |
| 20 | Pef1 | 3361 | 0.343 | 0.2154 | No |
| 21 | Sirt2 | 3751 | 0.318 | 0.2103 | No |
| 22 | Mmp15 | 4420 | 0.280 | 0.1920 | No |
| 23 | Vwf | 4430 | 0.280 | 0.2017 | No |
| 24 | Gda | 4557 | 0.273 | 0.2061 | No |
| 25 | F10 | 4759 | 0.262 | 0.2070 | No |
| 26 | Fn1 | 4991 | 0.251 | 0.2062 | No |
| 27 | Gsn | 5250 | 0.238 | 0.2038 | No |
| 28 | Serping1 | 5396 | 0.231 | 0.2060 | No |
| 29 | Tmprss6 | 5489 | 0.230 | 0.2103 | No |
| 30 | F9 | 5811 | 0.218 | 0.2045 | No |
| 31 | Timp1 | 6191 | 0.204 | 0.1957 | No |
| 32 | Klkb1 | 6306 | 0.202 | 0.1981 | No |
| 33 | Cfd | 6439 | 0.197 | 0.1996 | No |
| 34 | Hnf4a | 6521 | 0.194 | 0.2031 | No |
| 35 | Sparc | 6543 | 0.193 | 0.2092 | No |
| 36 | Pros1 | 6683 | 0.191 | 0.2101 | No |
| 37 | Lta4h | 6816 | 0.185 | 0.2112 | No |
| 38 | Wdr1 | 6928 | 0.180 | 0.2130 | No |
| 39 | Pf4 | 7000 | 0.178 | 0.2163 | No |
| 40 | Olr1 | 7387 | 0.163 | 0.2058 | No |
| 41 | Dct | 7727 | 0.151 | 0.1968 | No |
| 42 | C2 | 7765 | 0.150 | 0.2006 | No |
| 43 | Gp1ba | 7886 | 0.145 | 0.2007 | No |
| 44 | Plg | 8006 | 0.140 | 0.2007 | No |
| 45 | Ctsh | 8064 | 0.137 | 0.2032 | No |
| 46 | Cfb | 8153 | 0.133 | 0.2043 | No |
| 47 | Mmp8 | 8491 | 0.120 | 0.1943 | No |
| 48 | Klf7 | 9105 | 0.097 | 0.1717 | No |
| 49 | S100a1 | 9152 | 0.096 | 0.1732 | No |
| 50 | Lamp2 | 9433 | 0.087 | 0.1644 | No |
| 51 | Thbs1 | 9738 | 0.076 | 0.1542 | No |
| 52 | Capn2 | 9951 | 0.068 | 0.1477 | No |
| 53 | Hmgcs2 | 10039 | 0.065 | 0.1463 | No |
| 54 | Apoc1 | 10161 | 0.060 | 0.1433 | No |
| 55 | Prep | 10217 | 0.058 | 0.1431 | No |
| 56 | Timp3 | 10219 | 0.058 | 0.1451 | No |
| 57 | C8g | 10444 | 0.050 | 0.1374 | No |
| 58 | Cfh | 10658 | 0.042 | 0.1299 | No |
| 59 | Cpn1 | 10684 | 0.041 | 0.1303 | No |
| 60 | C8a | 11029 | 0.029 | 0.1167 | No |
| 61 | Furin | 11818 | 0.004 | 0.0833 | No |
| 62 | Plek | 12412 | -0.008 | 0.0583 | No |
| 63 | Cd9 | 12962 | -0.027 | 0.0360 | No |
| 64 | Arf4 | 13111 | -0.032 | 0.0308 | No |
| 65 | F8 | 13429 | -0.044 | 0.0189 | No |
| 66 | Masp2 | 13764 | -0.055 | 0.0067 | No |
| 67 | Ctsk | 13999 | -0.063 | -0.0010 | No |
| 68 | Gng12 | 14041 | -0.064 | -0.0005 | No |
| 69 | Adam9 | 14427 | -0.079 | -0.0140 | No |
| 70 | F2rl2 | 14730 | -0.089 | -0.0236 | No |
| 71 | F12 | 14805 | -0.092 | -0.0235 | No |
| 72 | Sh2b2 | 14962 | -0.097 | -0.0266 | No |
| 73 | Pecam1 | 14982 | -0.098 | -0.0239 | No |
| 74 | Lgmn | 15362 | -0.112 | -0.0360 | No |
| 75 | Crip2 | 15370 | -0.112 | -0.0323 | No |
| 76 | Tfpi2 | 15371 | -0.112 | -0.0282 | No |
| 77 | Cpq | 15616 | -0.121 | -0.0342 | No |
| 78 | C9 | 16045 | -0.138 | -0.0475 | No |
| 79 | Fbn1 | 16201 | -0.144 | -0.0489 | No |
| 80 | Lrp1 | 16311 | -0.149 | -0.0482 | No |
| 81 | Casp9 | 16420 | -0.153 | -0.0473 | No |
| 82 | Thbd | 16508 | -0.157 | -0.0453 | No |
| 83 | Anxa1 | 16602 | -0.159 | -0.0435 | No |
| 84 | Mst1 | 16804 | -0.167 | -0.0461 | No |
| 85 | Trf | 16853 | -0.169 | -0.0420 | No |
| 86 | Mmp14 | 17211 | -0.181 | -0.0507 | No |
| 87 | Ctso | 17610 | -0.198 | -0.0605 | No |
| 88 | Rabif | 17678 | -0.201 | -0.0561 | No |
| 89 | F3 | 17706 | -0.202 | -0.0500 | No |
| 90 | Fyn | 17895 | -0.211 | -0.0504 | No |
| 91 | Cpb2 | 17917 | -0.212 | -0.0437 | No |
| 92 | Maff | 17984 | -0.215 | -0.0388 | No |
| 93 | Acox2 | 18038 | -0.217 | -0.0332 | No |
| 94 | Klk8 | 18307 | -0.225 | -0.0365 | No |
| 95 | Rapgef3 | 18417 | -0.231 | -0.0328 | No |
| 96 | Mmp2 | 19013 | -0.261 | -0.0487 | No |
| 97 | Ctsb | 19192 | -0.270 | -0.0466 | No |
| 98 | Capn5 | 19307 | -0.275 | -0.0416 | No |
| 99 | Pdgfb | 19332 | -0.276 | -0.0326 | No |
| 100 | Rac1 | 19852 | -0.303 | -0.0438 | No |
| 101 | Clu | 19888 | -0.306 | -0.0343 | No |
| 102 | Dpp4 | 20003 | -0.311 | -0.0280 | No |
| 103 | Msrb2 | 20145 | -0.316 | -0.0226 | No |
| 104 | Cfi | 20202 | -0.320 | -0.0135 | No |
| 105 | Iscu | 20256 | -0.323 | -0.0041 | No |
| 106 | Dusp14 | 20739 | -0.352 | -0.0119 | No |
| 107 | Csrp1 | 20816 | -0.358 | -0.0023 | No |
| 108 | Ctse | 21766 | -0.427 | -0.0273 | No |
| 109 | S100a13 | 22125 | -0.464 | -0.0258 | No |
| 110 | Ctsl | 22369 | -0.490 | -0.0185 | No |
| 111 | Bmp1 | 23139 | -0.621 | -0.0289 | No |
| 112 | Gnb2 | 23232 | -0.648 | -0.0095 | No |
| 113 | Mmp11 | 23412 | -0.726 | 0.0090 | No |