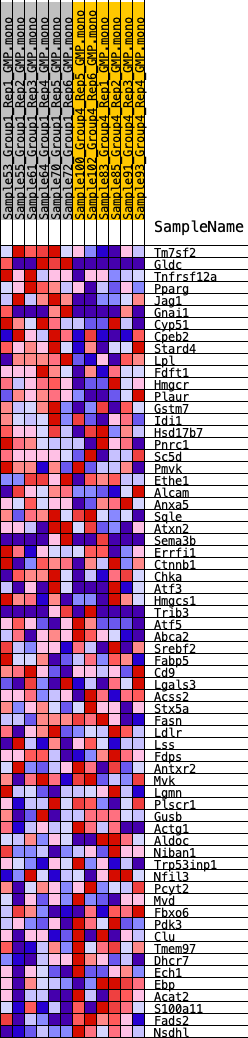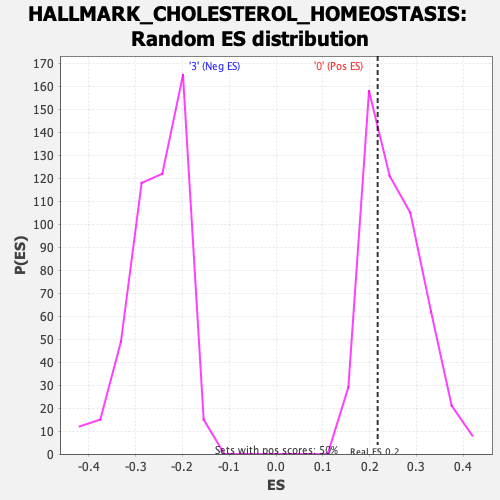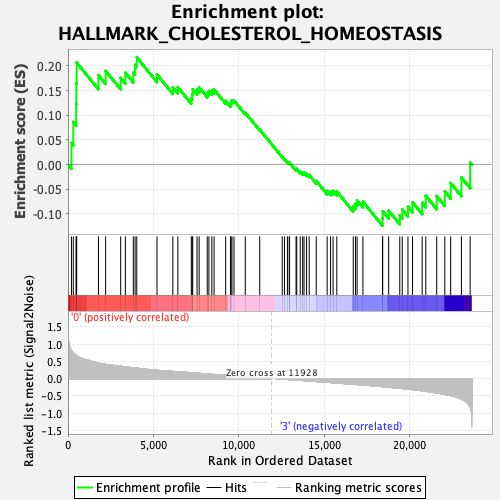
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group4.GMP.mono_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.21743773 |
| Normalized Enrichment Score (NES) | 0.85906845 |
| Nominal p-value | 0.64880955 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tm7sf2 | 202 | 0.808 | 0.0433 | Yes |
| 2 | Gldc | 310 | 0.741 | 0.0863 | Yes |
| 3 | Tnfrsf12a | 478 | 0.672 | 0.1224 | Yes |
| 4 | Pparg | 486 | 0.670 | 0.1652 | Yes |
| 5 | Jag1 | 508 | 0.660 | 0.2066 | Yes |
| 6 | Gnai1 | 1782 | 0.448 | 0.1814 | Yes |
| 7 | Cyp51 | 2201 | 0.412 | 0.1901 | Yes |
| 8 | Cpeb2 | 3083 | 0.359 | 0.1758 | Yes |
| 9 | Stard4 | 3356 | 0.343 | 0.1863 | Yes |
| 10 | Lpl | 3821 | 0.314 | 0.1867 | Yes |
| 11 | Fdft1 | 3924 | 0.307 | 0.2021 | Yes |
| 12 | Hmgcr | 4022 | 0.302 | 0.2174 | Yes |
| 13 | Plaur | 5206 | 0.240 | 0.1827 | No |
| 14 | Gstm7 | 6134 | 0.206 | 0.1566 | No |
| 15 | Idi1 | 6426 | 0.197 | 0.1569 | No |
| 16 | Hsd17b7 | 7215 | 0.170 | 0.1343 | No |
| 17 | Pnrc1 | 7264 | 0.168 | 0.1431 | No |
| 18 | Sc5d | 7296 | 0.166 | 0.1524 | No |
| 19 | Pmvk | 7550 | 0.158 | 0.1518 | No |
| 20 | Ethe1 | 7673 | 0.153 | 0.1565 | No |
| 21 | Alcam | 8147 | 0.133 | 0.1450 | No |
| 22 | Anxa5 | 8243 | 0.130 | 0.1493 | No |
| 23 | Sqle | 8415 | 0.123 | 0.1499 | No |
| 24 | Atxn2 | 8546 | 0.118 | 0.1520 | No |
| 25 | Sema3b | 9222 | 0.094 | 0.1293 | No |
| 26 | Errfi1 | 9507 | 0.084 | 0.1227 | No |
| 27 | Ctnnb1 | 9534 | 0.083 | 0.1269 | No |
| 28 | Chka | 9591 | 0.081 | 0.1297 | No |
| 29 | Atf3 | 9711 | 0.077 | 0.1297 | No |
| 30 | Hmgcs1 | 10371 | 0.053 | 0.1051 | No |
| 31 | Trib3 | 11216 | 0.023 | 0.0707 | No |
| 32 | Atf5 | 12533 | -0.012 | 0.0156 | No |
| 33 | Abca2 | 12662 | -0.017 | 0.0113 | No |
| 34 | Srebf2 | 12836 | -0.022 | 0.0054 | No |
| 35 | Fabp5 | 12941 | -0.026 | 0.0027 | No |
| 36 | Cd9 | 12962 | -0.027 | 0.0036 | No |
| 37 | Lgals3 | 13342 | -0.040 | -0.0099 | No |
| 38 | Acss2 | 13383 | -0.042 | -0.0090 | No |
| 39 | Stx5a | 13585 | -0.049 | -0.0144 | No |
| 40 | Fasn | 13726 | -0.053 | -0.0169 | No |
| 41 | Ldlr | 13808 | -0.056 | -0.0167 | No |
| 42 | Lss | 13957 | -0.061 | -0.0191 | No |
| 43 | Fdps | 14110 | -0.067 | -0.0212 | No |
| 44 | Antxr2 | 14522 | -0.083 | -0.0334 | No |
| 45 | Mvk | 15161 | -0.105 | -0.0537 | No |
| 46 | Lgmn | 15362 | -0.112 | -0.0550 | No |
| 47 | Plscr1 | 15509 | -0.117 | -0.0537 | No |
| 48 | Gusb | 15726 | -0.125 | -0.0548 | No |
| 49 | Actg1 | 16687 | -0.162 | -0.0852 | No |
| 50 | Aldoc | 16818 | -0.167 | -0.0799 | No |
| 51 | Niban1 | 16914 | -0.171 | -0.0730 | No |
| 52 | Trp53inp1 | 17254 | -0.183 | -0.0756 | No |
| 53 | Nfil3 | 18397 | -0.230 | -0.1093 | No |
| 54 | Pcyt2 | 18411 | -0.231 | -0.0950 | No |
| 55 | Mvd | 18760 | -0.247 | -0.0940 | No |
| 56 | Fbxo6 | 19410 | -0.280 | -0.1035 | No |
| 57 | Pdk3 | 19556 | -0.285 | -0.0914 | No |
| 58 | Clu | 19888 | -0.306 | -0.0858 | No |
| 59 | Tmem97 | 20156 | -0.317 | -0.0768 | No |
| 60 | Dhcr7 | 20720 | -0.351 | -0.0782 | No |
| 61 | Ech1 | 20931 | -0.366 | -0.0636 | No |
| 62 | Ebp | 21570 | -0.416 | -0.0640 | No |
| 63 | Acat2 | 22044 | -0.456 | -0.0548 | No |
| 64 | S100a11 | 22387 | -0.492 | -0.0377 | No |
| 65 | Fads2 | 23016 | -0.592 | -0.0264 | No |
| 66 | Nsdhl | 23526 | -0.812 | 0.0042 | No |