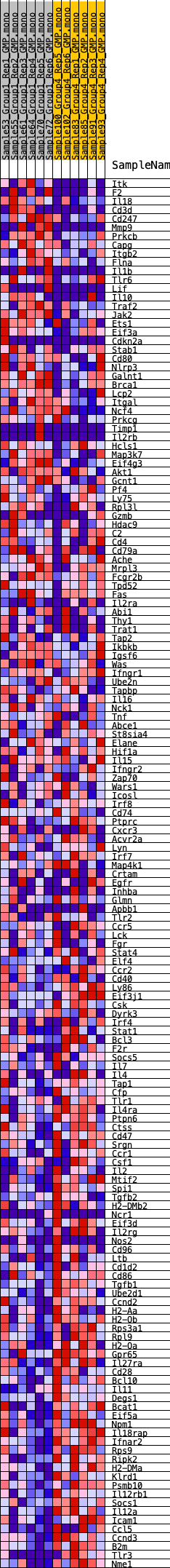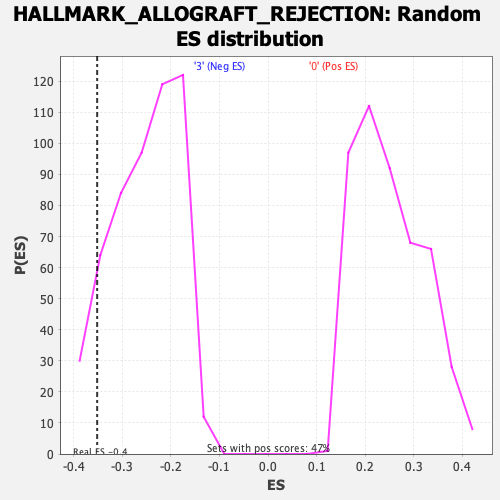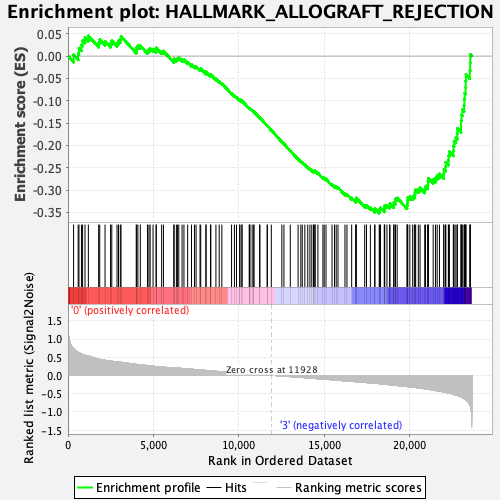
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group4.GMP.mono_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_ALLOGRAFT_REJECTION |
| Enrichment Score (ES) | -0.35169894 |
| Normalized Enrichment Score (NES) | -1.3973085 |
| Nominal p-value | 0.07954545 |
| FDR q-value | 0.28488857 |
| FWER p-Value | 0.606 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Itk | 326 | 0.735 | 0.0028 | No |
| 2 | F2 | 595 | 0.630 | 0.0056 | No |
| 3 | Il18 | 641 | 0.619 | 0.0178 | No |
| 4 | Cd3d | 787 | 0.587 | 0.0249 | No |
| 5 | Cd247 | 854 | 0.570 | 0.0350 | No |
| 6 | Mmp9 | 995 | 0.549 | 0.0415 | No |
| 7 | Prkcb | 1191 | 0.529 | 0.0452 | No |
| 8 | Capg | 1793 | 0.447 | 0.0297 | No |
| 9 | Itgb2 | 1855 | 0.439 | 0.0370 | No |
| 10 | Flna | 2167 | 0.414 | 0.0332 | No |
| 11 | Il1b | 2489 | 0.391 | 0.0284 | No |
| 12 | Tlr6 | 2562 | 0.388 | 0.0341 | No |
| 13 | Lif | 2868 | 0.370 | 0.0295 | No |
| 14 | Il10 | 2955 | 0.368 | 0.0341 | No |
| 15 | Traf2 | 3060 | 0.361 | 0.0379 | No |
| 16 | Jak2 | 3102 | 0.358 | 0.0443 | No |
| 17 | Ets1 | 3991 | 0.304 | 0.0133 | No |
| 18 | Eif3a | 4016 | 0.302 | 0.0192 | No |
| 19 | Cdkn2a | 4089 | 0.298 | 0.0228 | No |
| 20 | Stab1 | 4231 | 0.291 | 0.0234 | No |
| 21 | Cd80 | 4649 | 0.268 | 0.0117 | No |
| 22 | Nlrp3 | 4734 | 0.264 | 0.0141 | No |
| 23 | Galnt1 | 4805 | 0.260 | 0.0170 | No |
| 24 | Brca1 | 4976 | 0.252 | 0.0155 | No |
| 25 | Lcp2 | 5157 | 0.243 | 0.0133 | No |
| 26 | Itgal | 5163 | 0.243 | 0.0186 | No |
| 27 | Ncf4 | 5478 | 0.230 | 0.0105 | No |
| 28 | Prkcg | 5583 | 0.228 | 0.0112 | No |
| 29 | Timp1 | 6191 | 0.204 | -0.0100 | No |
| 30 | Il2rb | 6208 | 0.204 | -0.0061 | No |
| 31 | Hcls1 | 6341 | 0.200 | -0.0072 | No |
| 32 | Map3k7 | 6410 | 0.198 | -0.0056 | No |
| 33 | Eif4g3 | 6482 | 0.195 | -0.0042 | No |
| 34 | Akt1 | 6674 | 0.191 | -0.0080 | No |
| 35 | Gcnt1 | 6784 | 0.187 | -0.0084 | No |
| 36 | Pf4 | 7000 | 0.178 | -0.0135 | No |
| 37 | Ly75 | 7225 | 0.170 | -0.0192 | No |
| 38 | Rpl3l | 7398 | 0.162 | -0.0229 | No |
| 39 | Gzmb | 7493 | 0.159 | -0.0233 | No |
| 40 | Hdac9 | 7724 | 0.151 | -0.0296 | No |
| 41 | C2 | 7765 | 0.150 | -0.0279 | No |
| 42 | Cd4 | 8070 | 0.137 | -0.0378 | No |
| 43 | Cd79a | 8074 | 0.137 | -0.0348 | No |
| 44 | Ache | 8336 | 0.126 | -0.0431 | No |
| 45 | Mrpl3 | 8358 | 0.126 | -0.0411 | No |
| 46 | Fcgr2b | 8654 | 0.113 | -0.0511 | No |
| 47 | Tpd52 | 8845 | 0.107 | -0.0568 | No |
| 48 | Fas | 9000 | 0.102 | -0.0611 | No |
| 49 | Il2ra | 9569 | 0.082 | -0.0834 | No |
| 50 | Abi1 | 9736 | 0.076 | -0.0888 | No |
| 51 | Thy1 | 9857 | 0.072 | -0.0922 | No |
| 52 | Trat1 | 10046 | 0.064 | -0.0988 | No |
| 53 | Tap2 | 10055 | 0.064 | -0.0977 | No |
| 54 | Ikbkb | 10174 | 0.060 | -0.1014 | No |
| 55 | Igsf6 | 10182 | 0.060 | -0.1003 | No |
| 56 | Was | 10607 | 0.044 | -0.1174 | No |
| 57 | Ifngr1 | 10611 | 0.044 | -0.1165 | No |
| 58 | Ube2n | 10648 | 0.042 | -0.1171 | No |
| 59 | Tapbp | 10774 | 0.037 | -0.1216 | No |
| 60 | Il16 | 10873 | 0.034 | -0.1250 | No |
| 61 | Nck1 | 10877 | 0.034 | -0.1244 | No |
| 62 | Tnf | 11211 | 0.023 | -0.1380 | No |
| 63 | Abce1 | 11226 | 0.023 | -0.1381 | No |
| 64 | St8sia4 | 11653 | 0.009 | -0.1560 | No |
| 65 | Elane | 11661 | 0.009 | -0.1561 | No |
| 66 | Hif1a | 11894 | 0.001 | -0.1660 | No |
| 67 | Il15 | 12510 | -0.011 | -0.1919 | No |
| 68 | Ifngr2 | 12632 | -0.015 | -0.1967 | No |
| 69 | Zap70 | 13004 | -0.029 | -0.2119 | No |
| 70 | Wars1 | 13462 | -0.045 | -0.2304 | No |
| 71 | Icosl | 13622 | -0.051 | -0.2360 | No |
| 72 | Irf8 | 13704 | -0.052 | -0.2383 | No |
| 73 | Cd74 | 13856 | -0.058 | -0.2434 | No |
| 74 | Ptprc | 14027 | -0.064 | -0.2492 | No |
| 75 | Cxcr3 | 14154 | -0.069 | -0.2530 | No |
| 76 | Acvr2a | 14263 | -0.073 | -0.2560 | No |
| 77 | Lyn | 14365 | -0.077 | -0.2585 | No |
| 78 | Irf7 | 14388 | -0.078 | -0.2577 | No |
| 79 | Map4k1 | 14450 | -0.080 | -0.2585 | No |
| 80 | Crtam | 14452 | -0.080 | -0.2567 | No |
| 81 | Egfr | 14620 | -0.085 | -0.2619 | No |
| 82 | Inhba | 14913 | -0.095 | -0.2722 | No |
| 83 | Glmn | 14988 | -0.098 | -0.2731 | No |
| 84 | Apbb1 | 15094 | -0.102 | -0.2753 | No |
| 85 | Tlr2 | 15447 | -0.115 | -0.2877 | No |
| 86 | Ccr5 | 15591 | -0.120 | -0.2911 | No |
| 87 | Lck | 15689 | -0.124 | -0.2924 | No |
| 88 | Fgr | 15795 | -0.128 | -0.2940 | No |
| 89 | Stat4 | 16203 | -0.144 | -0.3080 | No |
| 90 | Elf4 | 16320 | -0.149 | -0.3096 | No |
| 91 | Ccr2 | 16595 | -0.159 | -0.3177 | No |
| 92 | Cd40 | 16836 | -0.168 | -0.3241 | No |
| 93 | Ly86 | 16850 | -0.169 | -0.3208 | No |
| 94 | Eif3j1 | 16863 | -0.169 | -0.3175 | No |
| 95 | Csk | 17358 | -0.188 | -0.3343 | No |
| 96 | Dyrk3 | 17462 | -0.192 | -0.3343 | No |
| 97 | Irf4 | 17690 | -0.202 | -0.3394 | No |
| 98 | Stat1 | 17942 | -0.212 | -0.3453 | No |
| 99 | Bcl3 | 17956 | -0.213 | -0.3410 | No |
| 100 | F2r | 18208 | -0.221 | -0.3467 | Yes |
| 101 | Socs5 | 18231 | -0.222 | -0.3426 | Yes |
| 102 | Il7 | 18284 | -0.224 | -0.3397 | Yes |
| 103 | Il4 | 18502 | -0.234 | -0.3437 | Yes |
| 104 | Tap1 | 18516 | -0.234 | -0.3389 | Yes |
| 105 | Cfp | 18531 | -0.235 | -0.3342 | Yes |
| 106 | Tlr1 | 18660 | -0.241 | -0.3342 | Yes |
| 107 | Il4ra | 18815 | -0.250 | -0.3351 | Yes |
| 108 | Ptpn6 | 18836 | -0.252 | -0.3302 | Yes |
| 109 | Ctss | 19058 | -0.264 | -0.3336 | Yes |
| 110 | Cd47 | 19064 | -0.264 | -0.3279 | Yes |
| 111 | Srgn | 19149 | -0.268 | -0.3254 | Yes |
| 112 | Ccr1 | 19153 | -0.268 | -0.3194 | Yes |
| 113 | Csf1 | 19257 | -0.272 | -0.3176 | Yes |
| 114 | Il2 | 19830 | -0.301 | -0.3352 | Yes |
| 115 | Mtif2 | 19856 | -0.303 | -0.3294 | Yes |
| 116 | Spi1 | 19863 | -0.303 | -0.3228 | Yes |
| 117 | Tgfb2 | 19879 | -0.305 | -0.3165 | Yes |
| 118 | H2-DMb2 | 19998 | -0.310 | -0.3145 | Yes |
| 119 | Ncr1 | 20165 | -0.317 | -0.3144 | Yes |
| 120 | Eif3d | 20263 | -0.323 | -0.3112 | Yes |
| 121 | Il2rg | 20300 | -0.325 | -0.3053 | Yes |
| 122 | Nos2 | 20340 | -0.327 | -0.2996 | Yes |
| 123 | Cd96 | 20497 | -0.337 | -0.2986 | Yes |
| 124 | Ltb | 20596 | -0.343 | -0.2950 | Yes |
| 125 | Cd1d2 | 20870 | -0.362 | -0.2984 | Yes |
| 126 | Cd86 | 20917 | -0.365 | -0.2921 | Yes |
| 127 | Tgfb1 | 21043 | -0.373 | -0.2890 | Yes |
| 128 | Ube2d1 | 21062 | -0.374 | -0.2813 | Yes |
| 129 | Ccnd2 | 21087 | -0.376 | -0.2738 | Yes |
| 130 | H2-Aa | 21364 | -0.398 | -0.2765 | Yes |
| 131 | H2-Ob | 21498 | -0.409 | -0.2729 | Yes |
| 132 | Rps3a1 | 21597 | -0.419 | -0.2676 | Yes |
| 133 | Rpl9 | 21730 | -0.425 | -0.2636 | Yes |
| 134 | H2-Oa | 21982 | -0.451 | -0.2640 | Yes |
| 135 | Gpr65 | 21989 | -0.452 | -0.2540 | Yes |
| 136 | Il27ra | 22090 | -0.461 | -0.2478 | Yes |
| 137 | Cd28 | 22102 | -0.462 | -0.2378 | Yes |
| 138 | Bcl10 | 22252 | -0.480 | -0.2333 | Yes |
| 139 | Il11 | 22267 | -0.481 | -0.2230 | Yes |
| 140 | Degs1 | 22313 | -0.485 | -0.2139 | Yes |
| 141 | Bcat1 | 22543 | -0.511 | -0.2121 | Yes |
| 142 | Eif5a | 22554 | -0.512 | -0.2009 | Yes |
| 143 | Npm1 | 22588 | -0.517 | -0.1906 | Yes |
| 144 | Il18rap | 22691 | -0.530 | -0.1829 | Yes |
| 145 | Ifnar2 | 22770 | -0.541 | -0.1740 | Yes |
| 146 | Rps9 | 22778 | -0.542 | -0.1620 | Yes |
| 147 | Ripk2 | 22993 | -0.586 | -0.1578 | Yes |
| 148 | H2-DMa | 22995 | -0.586 | -0.1445 | Yes |
| 149 | Klrd1 | 23024 | -0.593 | -0.1323 | Yes |
| 150 | Psmb10 | 23068 | -0.603 | -0.1204 | Yes |
| 151 | Il12rb1 | 23172 | -0.632 | -0.1105 | Yes |
| 152 | Socs1 | 23187 | -0.635 | -0.0967 | Yes |
| 153 | Il12a | 23209 | -0.640 | -0.0831 | Yes |
| 154 | Icam1 | 23259 | -0.657 | -0.0703 | Yes |
| 155 | Ccl5 | 23266 | -0.660 | -0.0556 | Yes |
| 156 | Ccnd3 | 23283 | -0.667 | -0.0411 | Yes |
| 157 | B2m | 23509 | -0.800 | -0.0326 | Yes |
| 158 | Tlr3 | 23528 | -0.819 | -0.0148 | Yes |
| 159 | Nme1 | 23537 | -0.831 | 0.0037 | Yes |