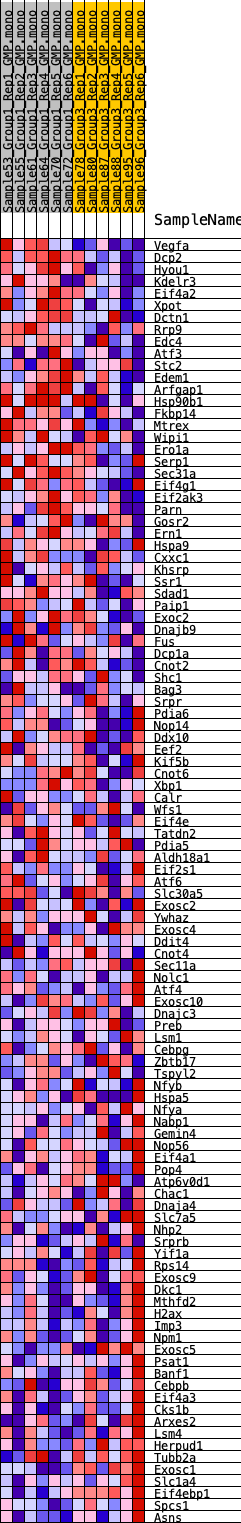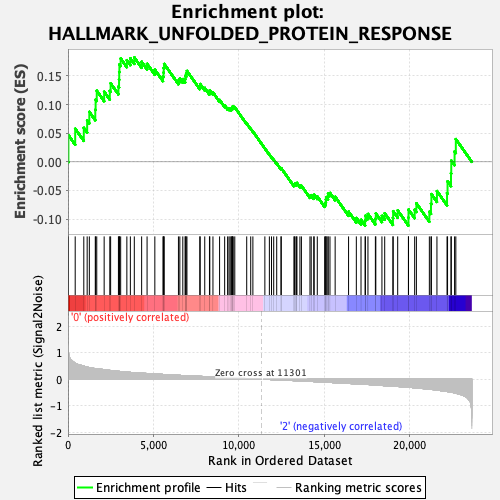
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group3.GMP.mono_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.18247794 |
| Normalized Enrichment Score (NES) | 0.7415564 |
| Nominal p-value | 0.7560484 |
| FDR q-value | 0.8245141 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Vegfa | 24 | 1.024 | 0.0467 | Yes |
| 2 | Dcp2 | 421 | 0.603 | 0.0580 | Yes |
| 3 | Hyou1 | 924 | 0.490 | 0.0594 | Yes |
| 4 | Kdelr3 | 1121 | 0.457 | 0.0724 | Yes |
| 5 | Eif4a2 | 1248 | 0.436 | 0.0874 | Yes |
| 6 | Xpot | 1596 | 0.398 | 0.0912 | Yes |
| 7 | Dctn1 | 1616 | 0.396 | 0.1088 | Yes |
| 8 | Rrp9 | 1682 | 0.391 | 0.1243 | Yes |
| 9 | Edc4 | 2115 | 0.360 | 0.1227 | Yes |
| 10 | Atf3 | 2446 | 0.331 | 0.1241 | Yes |
| 11 | Stc2 | 2494 | 0.326 | 0.1373 | Yes |
| 12 | Edem1 | 2950 | 0.295 | 0.1317 | Yes |
| 13 | Arfgap1 | 2987 | 0.294 | 0.1438 | Yes |
| 14 | Hsp90b1 | 2997 | 0.293 | 0.1571 | Yes |
| 15 | Fkbp14 | 3008 | 0.293 | 0.1703 | Yes |
| 16 | Mtrex | 3081 | 0.287 | 0.1806 | Yes |
| 17 | Wipi1 | 3443 | 0.266 | 0.1777 | Yes |
| 18 | Ero1a | 3648 | 0.255 | 0.1809 | Yes |
| 19 | Serp1 | 3879 | 0.244 | 0.1825 | Yes |
| 20 | Sec31a | 4308 | 0.227 | 0.1749 | No |
| 21 | Eif4g1 | 4626 | 0.213 | 0.1713 | No |
| 22 | Eif2ak3 | 5080 | 0.195 | 0.1611 | No |
| 23 | Parn | 5556 | 0.178 | 0.1492 | No |
| 24 | Gosr2 | 5590 | 0.177 | 0.1561 | No |
| 25 | Ern1 | 5599 | 0.177 | 0.1640 | No |
| 26 | Hspa9 | 5626 | 0.176 | 0.1711 | No |
| 27 | Cxxc1 | 6461 | 0.145 | 0.1424 | No |
| 28 | Khsrp | 6543 | 0.143 | 0.1456 | No |
| 29 | Ssr1 | 6716 | 0.136 | 0.1446 | No |
| 30 | Sdad1 | 6854 | 0.132 | 0.1450 | No |
| 31 | Paip1 | 6865 | 0.132 | 0.1507 | No |
| 32 | Exoc2 | 6915 | 0.130 | 0.1547 | No |
| 33 | Dnajb9 | 6958 | 0.129 | 0.1589 | No |
| 34 | Fus | 7711 | 0.107 | 0.1319 | No |
| 35 | Dcp1a | 7732 | 0.106 | 0.1360 | No |
| 36 | Cnot2 | 8000 | 0.098 | 0.1292 | No |
| 37 | Shc1 | 8276 | 0.090 | 0.1217 | No |
| 38 | Bag3 | 8296 | 0.090 | 0.1251 | No |
| 39 | Srpr | 8480 | 0.084 | 0.1212 | No |
| 40 | Pdia6 | 8868 | 0.072 | 0.1081 | No |
| 41 | Nop14 | 9165 | 0.064 | 0.0985 | No |
| 42 | Ddx10 | 9337 | 0.059 | 0.0940 | No |
| 43 | Eef2 | 9418 | 0.056 | 0.0932 | No |
| 44 | Kif5b | 9487 | 0.054 | 0.0928 | No |
| 45 | Cnot6 | 9556 | 0.052 | 0.0923 | No |
| 46 | Xbp1 | 9594 | 0.051 | 0.0931 | No |
| 47 | Calr | 9614 | 0.050 | 0.0946 | No |
| 48 | Wfs1 | 9622 | 0.050 | 0.0966 | No |
| 49 | Eif4e | 9667 | 0.048 | 0.0970 | No |
| 50 | Tatdn2 | 9757 | 0.045 | 0.0953 | No |
| 51 | Pdia5 | 10455 | 0.024 | 0.0668 | No |
| 52 | Aldh18a1 | 10689 | 0.018 | 0.0578 | No |
| 53 | Eif2s1 | 10816 | 0.014 | 0.0531 | No |
| 54 | Atf6 | 11517 | -0.002 | 0.0234 | No |
| 55 | Slc30a5 | 11781 | -0.011 | 0.0127 | No |
| 56 | Exosc2 | 11906 | -0.014 | 0.0081 | No |
| 57 | Ywhaz | 12034 | -0.018 | 0.0036 | No |
| 58 | Exosc4 | 12210 | -0.023 | -0.0028 | No |
| 59 | Ddit4 | 12460 | -0.031 | -0.0119 | No |
| 60 | Cnot4 | 12476 | -0.031 | -0.0111 | No |
| 61 | Sec11a | 13219 | -0.053 | -0.0402 | No |
| 62 | Nolc1 | 13246 | -0.054 | -0.0388 | No |
| 63 | Atf4 | 13286 | -0.055 | -0.0379 | No |
| 64 | Exosc10 | 13374 | -0.058 | -0.0389 | No |
| 65 | Dnajc3 | 13390 | -0.059 | -0.0368 | No |
| 66 | Preb | 13568 | -0.064 | -0.0413 | No |
| 67 | Lsm1 | 13655 | -0.067 | -0.0418 | No |
| 68 | Cebpg | 14156 | -0.082 | -0.0592 | No |
| 69 | Zbtb17 | 14228 | -0.085 | -0.0583 | No |
| 70 | Tspyl2 | 14379 | -0.089 | -0.0605 | No |
| 71 | Nfyb | 14400 | -0.090 | -0.0572 | No |
| 72 | Hspa5 | 14584 | -0.096 | -0.0605 | No |
| 73 | Nfya | 15014 | -0.110 | -0.0736 | No |
| 74 | Nabp1 | 15076 | -0.112 | -0.0710 | No |
| 75 | Gemin4 | 15088 | -0.113 | -0.0662 | No |
| 76 | Nop56 | 15103 | -0.113 | -0.0615 | No |
| 77 | Eif4a1 | 15210 | -0.117 | -0.0606 | No |
| 78 | Pop4 | 15216 | -0.117 | -0.0553 | No |
| 79 | Atp6v0d1 | 15326 | -0.121 | -0.0543 | No |
| 80 | Chac1 | 15629 | -0.132 | -0.0611 | No |
| 81 | Dnaja4 | 16414 | -0.159 | -0.0870 | No |
| 82 | Slc7a5 | 16868 | -0.174 | -0.0981 | No |
| 83 | Nhp2 | 17142 | -0.183 | -0.1012 | No |
| 84 | Srprb | 17382 | -0.193 | -0.1024 | No |
| 85 | Yif1a | 17399 | -0.193 | -0.0940 | No |
| 86 | Rps14 | 17548 | -0.199 | -0.0910 | No |
| 87 | Exosc9 | 17982 | -0.217 | -0.0993 | No |
| 88 | Dkc1 | 18008 | -0.219 | -0.0902 | No |
| 89 | Mthfd2 | 18366 | -0.236 | -0.0943 | No |
| 90 | H2ax | 18528 | -0.243 | -0.0899 | No |
| 91 | Imp3 | 19009 | -0.263 | -0.0980 | No |
| 92 | Npm1 | 19022 | -0.263 | -0.0863 | No |
| 93 | Exosc5 | 19288 | -0.273 | -0.0848 | No |
| 94 | Psat1 | 19915 | -0.303 | -0.0973 | No |
| 95 | Banf1 | 19921 | -0.304 | -0.0833 | No |
| 96 | Cebpb | 20281 | -0.323 | -0.0835 | No |
| 97 | Eif4a3 | 20381 | -0.328 | -0.0724 | No |
| 98 | Cks1b | 21140 | -0.377 | -0.0871 | No |
| 99 | Arxes2 | 21236 | -0.382 | -0.0733 | No |
| 100 | Lsm4 | 21260 | -0.384 | -0.0564 | No |
| 101 | Herpud1 | 21584 | -0.409 | -0.0510 | No |
| 102 | Tubb2a | 22175 | -0.462 | -0.0546 | No |
| 103 | Exosc1 | 22205 | -0.466 | -0.0341 | No |
| 104 | Slc1a4 | 22407 | -0.488 | -0.0199 | No |
| 105 | Eif4ebp1 | 22422 | -0.489 | 0.0023 | No |
| 106 | Spcs1 | 22616 | -0.517 | 0.0181 | No |
| 107 | Asns | 22689 | -0.529 | 0.0398 | No |