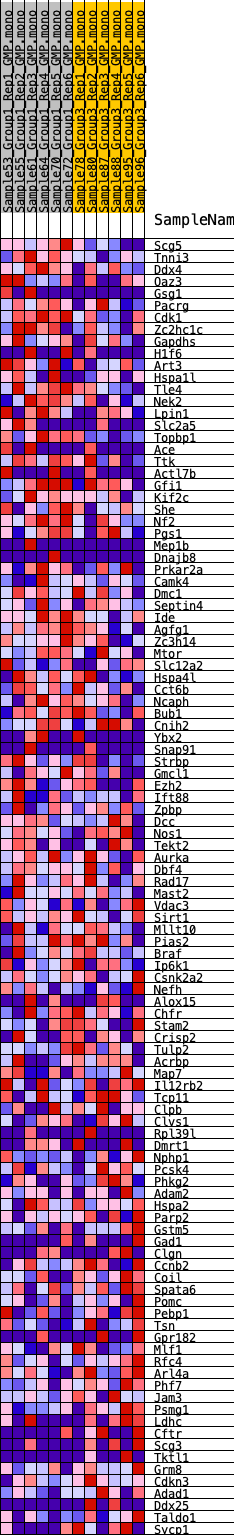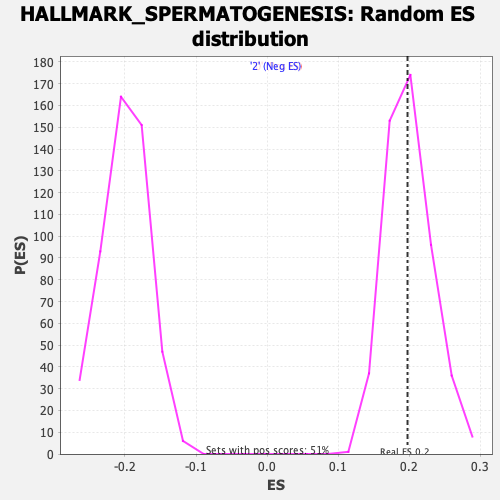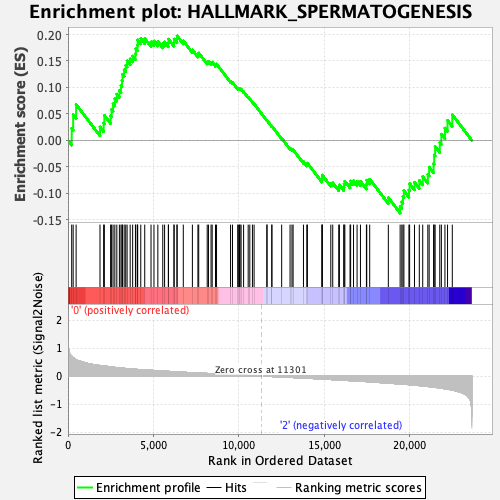
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group3.GMP.mono_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.19749555 |
| Normalized Enrichment Score (NES) | 0.9908059 |
| Nominal p-value | 0.48910892 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.986 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Scg5 | 216 | 0.708 | 0.0228 | Yes |
| 2 | Tnni3 | 302 | 0.658 | 0.0489 | Yes |
| 3 | Ddx4 | 475 | 0.581 | 0.0678 | Yes |
| 4 | Oaz3 | 1872 | 0.376 | 0.0254 | Yes |
| 5 | Gsg1 | 2084 | 0.361 | 0.0328 | Yes |
| 6 | Pacrg | 2127 | 0.359 | 0.0472 | Yes |
| 7 | Cdk1 | 2492 | 0.326 | 0.0464 | Yes |
| 8 | Zc2hc1c | 2551 | 0.322 | 0.0585 | Yes |
| 9 | Gapdhs | 2636 | 0.317 | 0.0692 | Yes |
| 10 | H1f6 | 2740 | 0.309 | 0.0788 | Yes |
| 11 | Art3 | 2856 | 0.301 | 0.0875 | Yes |
| 12 | Hspa1l | 3006 | 0.293 | 0.0943 | Yes |
| 13 | Tle4 | 3099 | 0.286 | 0.1033 | Yes |
| 14 | Nek2 | 3159 | 0.282 | 0.1136 | Yes |
| 15 | Lpin1 | 3190 | 0.280 | 0.1250 | Yes |
| 16 | Slc2a5 | 3286 | 0.275 | 0.1333 | Yes |
| 17 | Topbp1 | 3377 | 0.270 | 0.1417 | Yes |
| 18 | Ace | 3457 | 0.265 | 0.1503 | Yes |
| 19 | Ttk | 3637 | 0.256 | 0.1542 | Yes |
| 20 | Actl7b | 3782 | 0.249 | 0.1593 | Yes |
| 21 | Gfi1 | 3945 | 0.241 | 0.1633 | Yes |
| 22 | Kif2c | 3968 | 0.240 | 0.1732 | Yes |
| 23 | She | 4065 | 0.235 | 0.1798 | Yes |
| 24 | Nf2 | 4077 | 0.234 | 0.1898 | Yes |
| 25 | Pgs1 | 4257 | 0.229 | 0.1926 | Yes |
| 26 | Mep1b | 4489 | 0.218 | 0.1926 | Yes |
| 27 | Dnajb8 | 4857 | 0.204 | 0.1862 | Yes |
| 28 | Prkar2a | 5029 | 0.197 | 0.1878 | Yes |
| 29 | Camk4 | 5256 | 0.190 | 0.1868 | Yes |
| 30 | Dmc1 | 5544 | 0.179 | 0.1827 | Yes |
| 31 | Septin4 | 5651 | 0.175 | 0.1861 | Yes |
| 32 | Ide | 5869 | 0.166 | 0.1844 | Yes |
| 33 | Agfg1 | 5880 | 0.166 | 0.1914 | Yes |
| 34 | Zc3h14 | 6200 | 0.154 | 0.1848 | Yes |
| 35 | Mtor | 6207 | 0.154 | 0.1915 | Yes |
| 36 | Slc12a2 | 6377 | 0.148 | 0.1910 | Yes |
| 37 | Hspa4l | 6382 | 0.148 | 0.1975 | Yes |
| 38 | Cct6b | 6745 | 0.136 | 0.1882 | No |
| 39 | Ncaph | 7277 | 0.118 | 0.1710 | No |
| 40 | Bub1 | 7601 | 0.110 | 0.1622 | No |
| 41 | Cnih2 | 7647 | 0.108 | 0.1652 | No |
| 42 | Ybx2 | 8144 | 0.093 | 0.1483 | No |
| 43 | Snap91 | 8217 | 0.092 | 0.1494 | No |
| 44 | Strbp | 8369 | 0.088 | 0.1469 | No |
| 45 | Gmcl1 | 8435 | 0.086 | 0.1480 | No |
| 46 | Ezh2 | 8629 | 0.080 | 0.1434 | No |
| 47 | Ift88 | 8690 | 0.078 | 0.1444 | No |
| 48 | Zpbp | 9510 | 0.053 | 0.1119 | No |
| 49 | Dcc | 9623 | 0.049 | 0.1094 | No |
| 50 | Nos1 | 9930 | 0.041 | 0.0982 | No |
| 51 | Tekt2 | 9964 | 0.040 | 0.0986 | No |
| 52 | Aurka | 10033 | 0.037 | 0.0974 | No |
| 53 | Dbf4 | 10063 | 0.036 | 0.0978 | No |
| 54 | Rad17 | 10126 | 0.035 | 0.0967 | No |
| 55 | Mast2 | 10273 | 0.030 | 0.0919 | No |
| 56 | Vdac3 | 10541 | 0.022 | 0.0815 | No |
| 57 | Sirt1 | 10636 | 0.019 | 0.0784 | No |
| 58 | Mllt10 | 10790 | 0.015 | 0.0725 | No |
| 59 | Pias2 | 10887 | 0.012 | 0.0690 | No |
| 60 | Braf | 11633 | -0.006 | 0.0376 | No |
| 61 | Ip6k1 | 11645 | -0.006 | 0.0374 | No |
| 62 | Csnk2a2 | 11925 | -0.015 | 0.0262 | No |
| 63 | Nefh | 11926 | -0.015 | 0.0269 | No |
| 64 | Alox15 | 12500 | -0.031 | 0.0040 | No |
| 65 | Chfr | 12991 | -0.046 | -0.0148 | No |
| 66 | Stam2 | 13103 | -0.049 | -0.0173 | No |
| 67 | Crisp2 | 13169 | -0.051 | -0.0177 | No |
| 68 | Tulp2 | 13775 | -0.071 | -0.0403 | No |
| 69 | Acrbp | 13972 | -0.076 | -0.0452 | No |
| 70 | Map7 | 14005 | -0.077 | -0.0430 | No |
| 71 | Il12rb2 | 14854 | -0.105 | -0.0744 | No |
| 72 | Tcp11 | 14870 | -0.106 | -0.0702 | No |
| 73 | Clpb | 14875 | -0.106 | -0.0656 | No |
| 74 | Clvs1 | 15375 | -0.123 | -0.0813 | No |
| 75 | Rpl39l | 15488 | -0.127 | -0.0803 | No |
| 76 | Dmrt1 | 15839 | -0.139 | -0.0889 | No |
| 77 | Nphp1 | 15878 | -0.140 | -0.0842 | No |
| 78 | Pcsk4 | 16133 | -0.149 | -0.0883 | No |
| 79 | Phkg2 | 16152 | -0.150 | -0.0823 | No |
| 80 | Adam2 | 16196 | -0.152 | -0.0772 | No |
| 81 | Hspa2 | 16501 | -0.162 | -0.0828 | No |
| 82 | Parp2 | 16535 | -0.163 | -0.0769 | No |
| 83 | Gstm5 | 16704 | -0.170 | -0.0763 | No |
| 84 | Gad1 | 16912 | -0.175 | -0.0772 | No |
| 85 | Clgn | 17109 | -0.182 | -0.0774 | No |
| 86 | Ccnb2 | 17458 | -0.195 | -0.0833 | No |
| 87 | Coil | 17477 | -0.196 | -0.0753 | No |
| 88 | Spata6 | 17650 | -0.203 | -0.0734 | No |
| 89 | Pomc | 18740 | -0.252 | -0.1083 | No |
| 90 | Pebp1 | 19431 | -0.280 | -0.1250 | No |
| 91 | Tsn | 19528 | -0.285 | -0.1162 | No |
| 92 | Gpr182 | 19596 | -0.288 | -0.1061 | No |
| 93 | Mlf1 | 19651 | -0.291 | -0.0952 | No |
| 94 | Rfc4 | 19946 | -0.305 | -0.0939 | No |
| 95 | Arl4a | 19986 | -0.307 | -0.0817 | No |
| 96 | Phf7 | 20270 | -0.323 | -0.0792 | No |
| 97 | Jam3 | 20552 | -0.338 | -0.0758 | No |
| 98 | Psmg1 | 20752 | -0.352 | -0.0684 | No |
| 99 | Ldhc | 21051 | -0.371 | -0.0643 | No |
| 100 | Cftr | 21128 | -0.376 | -0.0506 | No |
| 101 | Scg3 | 21388 | -0.393 | -0.0438 | No |
| 102 | Tktl1 | 21423 | -0.396 | -0.0274 | No |
| 103 | Grm8 | 21478 | -0.400 | -0.0117 | No |
| 104 | Cdkn3 | 21750 | -0.425 | -0.0040 | No |
| 105 | Adad1 | 21843 | -0.435 | 0.0117 | No |
| 106 | Ddx25 | 22052 | -0.452 | 0.0233 | No |
| 107 | Taldo1 | 22204 | -0.466 | 0.0379 | No |
| 108 | Sycp1 | 22482 | -0.497 | 0.0486 | No |