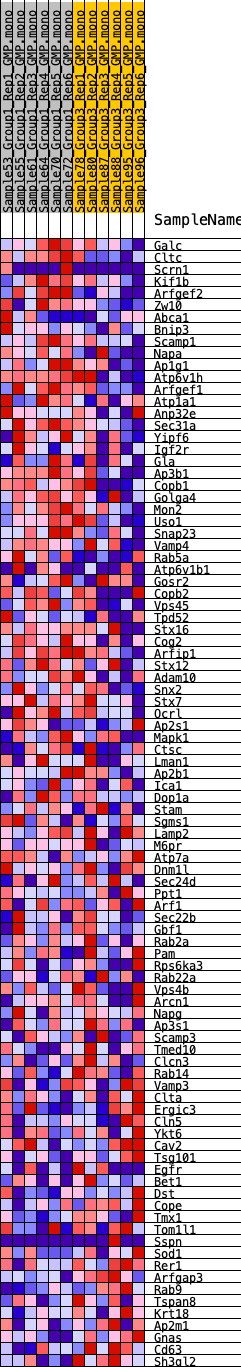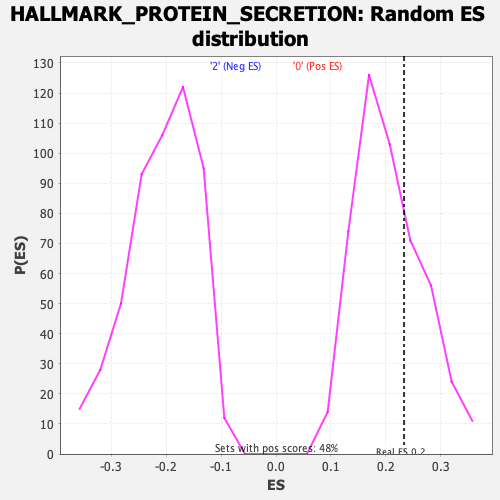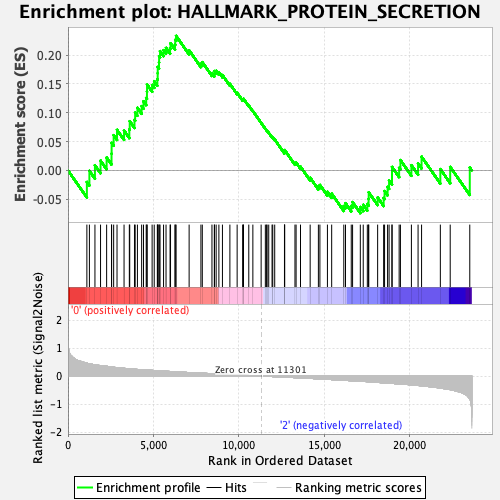
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group3.GMP.mono_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.23316285 |
| Normalized Enrichment Score (NES) | 1.1366495 |
| Nominal p-value | 0.2881002 |
| FDR q-value | 0.9947628 |
| FWER p-Value | 0.94 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Galc | 1104 | 0.460 | -0.0199 | Yes |
| 2 | Cltc | 1252 | 0.436 | -0.0006 | Yes |
| 3 | Scrn1 | 1576 | 0.399 | 0.0090 | Yes |
| 4 | Kif1b | 1904 | 0.373 | 0.0170 | Yes |
| 5 | Arfgef2 | 2260 | 0.348 | 0.0224 | Yes |
| 6 | Zw10 | 2546 | 0.322 | 0.0291 | Yes |
| 7 | Abca1 | 2552 | 0.321 | 0.0478 | Yes |
| 8 | Bnip3 | 2671 | 0.313 | 0.0611 | Yes |
| 9 | Scamp1 | 2868 | 0.300 | 0.0704 | Yes |
| 10 | Napa | 3279 | 0.276 | 0.0691 | Yes |
| 11 | Ap1g1 | 3588 | 0.258 | 0.0712 | Yes |
| 12 | Atp6v1h | 3611 | 0.257 | 0.0853 | Yes |
| 13 | Arfgef1 | 3889 | 0.244 | 0.0878 | Yes |
| 14 | Atp1a1 | 3929 | 0.242 | 0.1003 | Yes |
| 15 | Anp32e | 4061 | 0.235 | 0.1086 | Yes |
| 16 | Sec31a | 4308 | 0.227 | 0.1114 | Yes |
| 17 | Yipf6 | 4416 | 0.221 | 0.1198 | Yes |
| 18 | Igf2r | 4575 | 0.216 | 0.1258 | Yes |
| 19 | Gla | 4619 | 0.214 | 0.1365 | Yes |
| 20 | Ap3b1 | 4625 | 0.213 | 0.1488 | Yes |
| 21 | Copb1 | 4921 | 0.203 | 0.1481 | Yes |
| 22 | Golga4 | 5044 | 0.197 | 0.1545 | Yes |
| 23 | Mon2 | 5226 | 0.191 | 0.1580 | Yes |
| 24 | Uso1 | 5246 | 0.191 | 0.1684 | Yes |
| 25 | Snap23 | 5250 | 0.190 | 0.1794 | Yes |
| 26 | Vamp4 | 5324 | 0.187 | 0.1873 | Yes |
| 27 | Rab5a | 5331 | 0.187 | 0.1980 | Yes |
| 28 | Atp6v1b1 | 5384 | 0.185 | 0.2067 | Yes |
| 29 | Gosr2 | 5590 | 0.177 | 0.2084 | Yes |
| 30 | Copb2 | 5735 | 0.172 | 0.2123 | Yes |
| 31 | Vps45 | 5972 | 0.163 | 0.2119 | Yes |
| 32 | Tpd52 | 5998 | 0.162 | 0.2203 | Yes |
| 33 | Stx16 | 6260 | 0.152 | 0.2181 | Yes |
| 34 | Cog2 | 6274 | 0.152 | 0.2264 | Yes |
| 35 | Arfip1 | 6324 | 0.150 | 0.2332 | Yes |
| 36 | Stx12 | 7086 | 0.124 | 0.2081 | No |
| 37 | Adam10 | 7776 | 0.105 | 0.1850 | No |
| 38 | Snx2 | 7857 | 0.103 | 0.1876 | No |
| 39 | Stx7 | 8422 | 0.086 | 0.1687 | No |
| 40 | Ocrl | 8563 | 0.082 | 0.1676 | No |
| 41 | Ap2s1 | 8567 | 0.082 | 0.1722 | No |
| 42 | Mapk1 | 8667 | 0.078 | 0.1726 | No |
| 43 | Ctsc | 8827 | 0.073 | 0.1702 | No |
| 44 | Lman1 | 9035 | 0.067 | 0.1653 | No |
| 45 | Ap2b1 | 9471 | 0.054 | 0.1500 | No |
| 46 | Ica1 | 9895 | 0.042 | 0.1344 | No |
| 47 | Dop1a | 10218 | 0.031 | 0.1226 | No |
| 48 | Stam | 10255 | 0.030 | 0.1228 | No |
| 49 | Sgms1 | 10267 | 0.030 | 0.1241 | No |
| 50 | Lamp2 | 10576 | 0.021 | 0.1123 | No |
| 51 | M6pr | 10815 | 0.014 | 0.1030 | No |
| 52 | Atp7a | 11299 | 0.000 | 0.0825 | No |
| 53 | Dnm1l | 11545 | -0.003 | 0.0723 | No |
| 54 | Sec24d | 11619 | -0.006 | 0.0695 | No |
| 55 | Ppt1 | 11646 | -0.006 | 0.0688 | No |
| 56 | Arf1 | 11743 | -0.010 | 0.0653 | No |
| 57 | Sec22b | 11938 | -0.016 | 0.0579 | No |
| 58 | Gbf1 | 11979 | -0.017 | 0.0572 | No |
| 59 | Rab2a | 12093 | -0.020 | 0.0536 | No |
| 60 | Pam | 12664 | -0.036 | 0.0315 | No |
| 61 | Rps6ka3 | 12670 | -0.036 | 0.0334 | No |
| 62 | Rab22a | 12675 | -0.036 | 0.0353 | No |
| 63 | Vps4b | 13278 | -0.055 | 0.0130 | No |
| 64 | Arcn1 | 13344 | -0.057 | 0.0136 | No |
| 65 | Napg | 13602 | -0.065 | 0.0065 | No |
| 66 | Ap3s1 | 14167 | -0.083 | -0.0126 | No |
| 67 | Scamp3 | 14648 | -0.098 | -0.0273 | No |
| 68 | Tmed10 | 14737 | -0.101 | -0.0251 | No |
| 69 | Clcn3 | 15177 | -0.116 | -0.0370 | No |
| 70 | Rab14 | 15426 | -0.124 | -0.0402 | No |
| 71 | Vamp3 | 16135 | -0.150 | -0.0615 | No |
| 72 | Clta | 16234 | -0.153 | -0.0567 | No |
| 73 | Ergic3 | 16571 | -0.165 | -0.0613 | No |
| 74 | Cln5 | 16649 | -0.167 | -0.0548 | No |
| 75 | Ykt6 | 17101 | -0.181 | -0.0633 | No |
| 76 | Cav2 | 17272 | -0.188 | -0.0595 | No |
| 77 | Tsg101 | 17511 | -0.197 | -0.0580 | No |
| 78 | Egfr | 17585 | -0.200 | -0.0494 | No |
| 79 | Bet1 | 17593 | -0.201 | -0.0379 | No |
| 80 | Dst | 18115 | -0.225 | -0.0469 | No |
| 81 | Cope | 18466 | -0.241 | -0.0477 | No |
| 82 | Tmx1 | 18520 | -0.242 | -0.0357 | No |
| 83 | Tom1l1 | 18697 | -0.251 | -0.0285 | No |
| 84 | Sspn | 18784 | -0.254 | -0.0173 | No |
| 85 | Sod1 | 18955 | -0.261 | -0.0092 | No |
| 86 | Rer1 | 18956 | -0.261 | 0.0061 | No |
| 87 | Arfgap3 | 19373 | -0.277 | 0.0047 | No |
| 88 | Rab9 | 19448 | -0.281 | 0.0180 | No |
| 89 | Tspan8 | 20090 | -0.312 | 0.0090 | No |
| 90 | Krt18 | 20480 | -0.335 | 0.0121 | No |
| 91 | Ap2m1 | 20685 | -0.348 | 0.0238 | No |
| 92 | Gnas | 21783 | -0.428 | 0.0023 | No |
| 93 | Cd63 | 22360 | -0.482 | 0.0061 | No |
| 94 | Sh3gl2 | 23505 | -0.811 | 0.0051 | No |