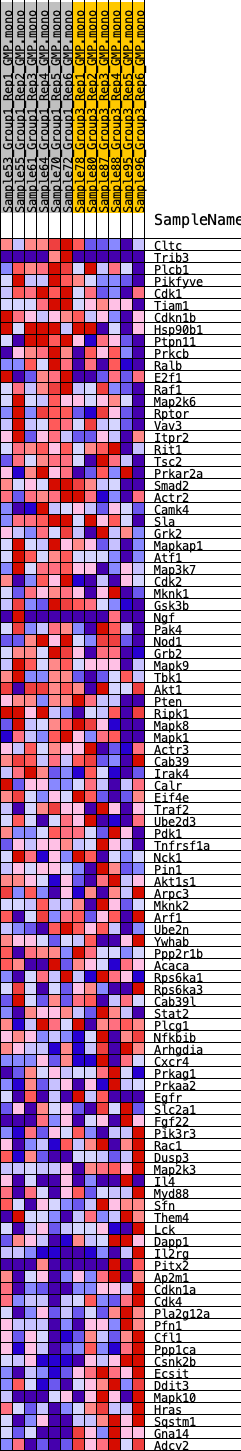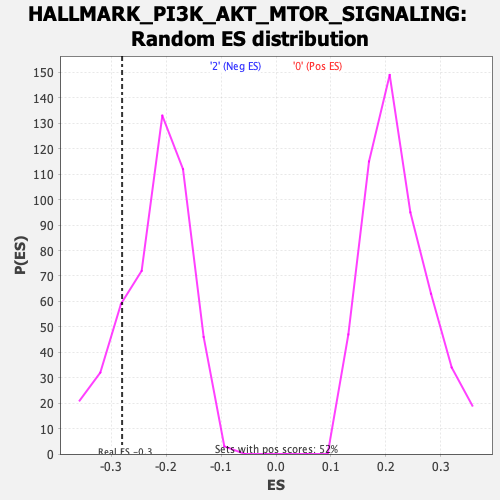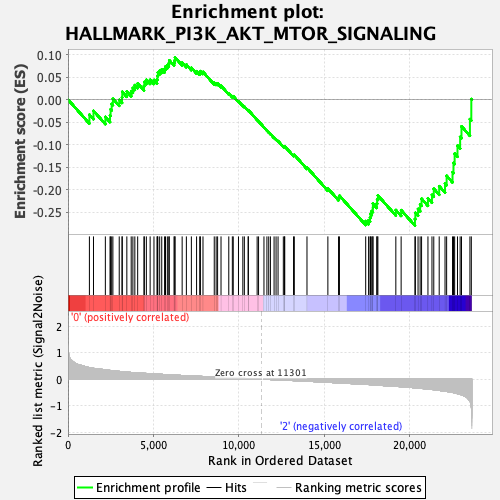
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group3.GMP.mono_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.27983823 |
| Normalized Enrichment Score (NES) | -1.282267 |
| Nominal p-value | 0.15062761 |
| FDR q-value | 0.6045744 |
| FWER p-Value | 0.798 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cltc | 1252 | 0.436 | -0.0334 | No |
| 2 | Trib3 | 1491 | 0.409 | -0.0250 | No |
| 3 | Plcb1 | 2185 | 0.353 | -0.0384 | No |
| 4 | Pikfyve | 2459 | 0.329 | -0.0351 | No |
| 5 | Cdk1 | 2492 | 0.326 | -0.0216 | No |
| 6 | Tiam1 | 2556 | 0.321 | -0.0097 | No |
| 7 | Cdkn1b | 2626 | 0.318 | 0.0018 | No |
| 8 | Hsp90b1 | 2997 | 0.293 | -0.0006 | No |
| 9 | Ptpn11 | 3161 | 0.282 | 0.0052 | No |
| 10 | Prkcb | 3174 | 0.281 | 0.0175 | No |
| 11 | Ralb | 3442 | 0.266 | 0.0182 | No |
| 12 | E2f1 | 3697 | 0.253 | 0.0189 | No |
| 13 | Raf1 | 3796 | 0.248 | 0.0260 | No |
| 14 | Map2k6 | 3910 | 0.243 | 0.0322 | No |
| 15 | Rptor | 4076 | 0.234 | 0.0358 | No |
| 16 | Vav3 | 4444 | 0.220 | 0.0302 | No |
| 17 | Itpr2 | 4466 | 0.219 | 0.0393 | No |
| 18 | Rit1 | 4584 | 0.215 | 0.0441 | No |
| 19 | Tsc2 | 4805 | 0.206 | 0.0441 | No |
| 20 | Prkar2a | 5029 | 0.197 | 0.0436 | No |
| 21 | Smad2 | 5208 | 0.192 | 0.0447 | No |
| 22 | Actr2 | 5234 | 0.191 | 0.0523 | No |
| 23 | Camk4 | 5256 | 0.190 | 0.0601 | No |
| 24 | Sla | 5363 | 0.186 | 0.0640 | No |
| 25 | Grk2 | 5484 | 0.181 | 0.0672 | No |
| 26 | Mapkap1 | 5650 | 0.175 | 0.0681 | No |
| 27 | Atf1 | 5704 | 0.173 | 0.0737 | No |
| 28 | Map3k7 | 5812 | 0.169 | 0.0768 | No |
| 29 | Cdk2 | 5897 | 0.165 | 0.0807 | No |
| 30 | Mknk1 | 5925 | 0.164 | 0.0871 | No |
| 31 | Gsk3b | 6203 | 0.154 | 0.0823 | No |
| 32 | Ngf | 6246 | 0.152 | 0.0874 | No |
| 33 | Pak4 | 6262 | 0.152 | 0.0937 | No |
| 34 | Nod1 | 6678 | 0.138 | 0.0823 | No |
| 35 | Grb2 | 6925 | 0.130 | 0.0777 | No |
| 36 | Mapk9 | 7217 | 0.120 | 0.0708 | No |
| 37 | Tbk1 | 7522 | 0.113 | 0.0630 | No |
| 38 | Akt1 | 7706 | 0.107 | 0.0601 | No |
| 39 | Pten | 7747 | 0.106 | 0.0632 | No |
| 40 | Ripk1 | 7897 | 0.101 | 0.0615 | No |
| 41 | Mapk8 | 8556 | 0.082 | 0.0372 | No |
| 42 | Mapk1 | 8667 | 0.078 | 0.0361 | No |
| 43 | Actr3 | 8752 | 0.076 | 0.0360 | No |
| 44 | Cab39 | 8952 | 0.069 | 0.0307 | No |
| 45 | Irak4 | 9406 | 0.057 | 0.0140 | No |
| 46 | Calr | 9614 | 0.050 | 0.0074 | No |
| 47 | Eif4e | 9667 | 0.048 | 0.0074 | No |
| 48 | Traf2 | 9973 | 0.039 | -0.0038 | No |
| 49 | Ube2d3 | 10228 | 0.031 | -0.0132 | No |
| 50 | Pdk1 | 10317 | 0.029 | -0.0156 | No |
| 51 | Tnfrsf1a | 10540 | 0.022 | -0.0241 | No |
| 52 | Nck1 | 10551 | 0.021 | -0.0235 | No |
| 53 | Pin1 | 11065 | 0.007 | -0.0450 | No |
| 54 | Akt1s1 | 11148 | 0.005 | -0.0483 | No |
| 55 | Arpc3 | 11464 | -0.001 | -0.0616 | No |
| 56 | Mknk2 | 11637 | -0.006 | -0.0686 | No |
| 57 | Arf1 | 11743 | -0.010 | -0.0727 | No |
| 58 | Ube2n | 11842 | -0.013 | -0.0763 | No |
| 59 | Ywhab | 12056 | -0.019 | -0.0845 | No |
| 60 | Ppp2r1b | 12172 | -0.022 | -0.0883 | No |
| 61 | Acaca | 12289 | -0.026 | -0.0921 | No |
| 62 | Rps6ka1 | 12597 | -0.034 | -0.1036 | No |
| 63 | Rps6ka3 | 12670 | -0.036 | -0.1050 | No |
| 64 | Cab39l | 12677 | -0.036 | -0.1036 | No |
| 65 | Stat2 | 13194 | -0.052 | -0.1232 | No |
| 66 | Plcg1 | 13240 | -0.054 | -0.1227 | No |
| 67 | Nfkbib | 13981 | -0.076 | -0.1507 | No |
| 68 | Arhgdia | 15203 | -0.117 | -0.1973 | No |
| 69 | Cxcr4 | 15827 | -0.138 | -0.2175 | No |
| 70 | Prkag1 | 15880 | -0.140 | -0.2133 | No |
| 71 | Prkaa2 | 17416 | -0.194 | -0.2698 | Yes |
| 72 | Egfr | 17585 | -0.200 | -0.2678 | Yes |
| 73 | Slc2a1 | 17670 | -0.204 | -0.2621 | Yes |
| 74 | Fgf22 | 17696 | -0.206 | -0.2538 | Yes |
| 75 | Pik3r3 | 17770 | -0.209 | -0.2475 | Yes |
| 76 | Rac1 | 17836 | -0.211 | -0.2406 | Yes |
| 77 | Dusp3 | 17838 | -0.211 | -0.2311 | Yes |
| 78 | Map2k3 | 18065 | -0.221 | -0.2307 | Yes |
| 79 | Il4 | 18086 | -0.223 | -0.2214 | Yes |
| 80 | Myd88 | 18129 | -0.225 | -0.2129 | Yes |
| 81 | Sfn | 19179 | -0.270 | -0.2453 | Yes |
| 82 | Them4 | 19489 | -0.283 | -0.2455 | Yes |
| 83 | Lck | 20297 | -0.324 | -0.2651 | Yes |
| 84 | Dapp1 | 20322 | -0.325 | -0.2514 | Yes |
| 85 | Il2rg | 20479 | -0.334 | -0.2428 | Yes |
| 86 | Pitx2 | 20601 | -0.341 | -0.2325 | Yes |
| 87 | Ap2m1 | 20685 | -0.348 | -0.2202 | Yes |
| 88 | Cdkn1a | 21058 | -0.372 | -0.2191 | Yes |
| 89 | Cdk4 | 21293 | -0.387 | -0.2115 | Yes |
| 90 | Pla2g12a | 21399 | -0.394 | -0.1981 | Yes |
| 91 | Pfn1 | 21716 | -0.421 | -0.1924 | Yes |
| 92 | Cfl1 | 22057 | -0.452 | -0.1863 | Yes |
| 93 | Ppp1ca | 22151 | -0.461 | -0.1693 | Yes |
| 94 | Csnk2b | 22496 | -0.499 | -0.1613 | Yes |
| 95 | Ecsit | 22552 | -0.506 | -0.1406 | Yes |
| 96 | Ddit3 | 22620 | -0.517 | -0.1200 | Yes |
| 97 | Mapk10 | 22786 | -0.547 | -0.1022 | Yes |
| 98 | Hras | 22941 | -0.571 | -0.0828 | Yes |
| 99 | Sqstm1 | 23020 | -0.586 | -0.0595 | Yes |
| 100 | Gna14 | 23514 | -0.818 | -0.0432 | Yes |
| 101 | Adcy2 | 23595 | -1.054 | 0.0012 | Yes |