Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group3.GMP.mono_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
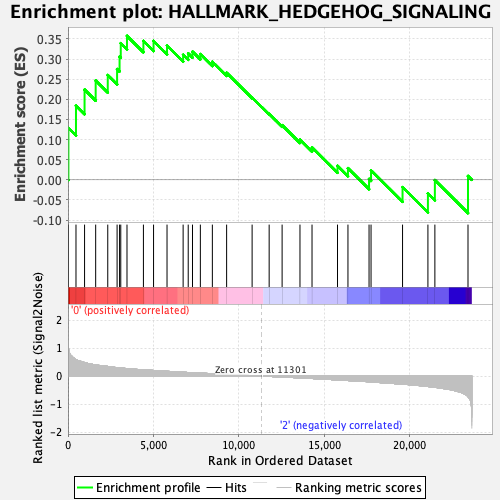
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
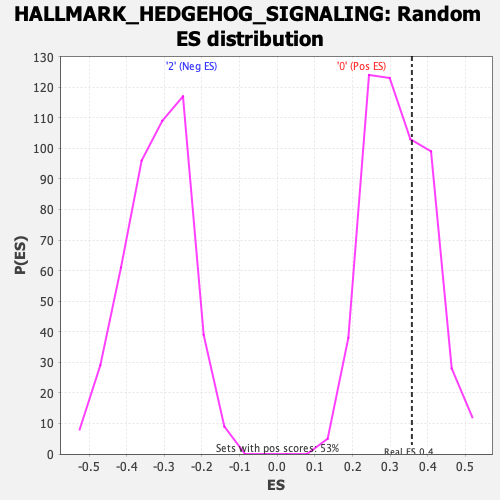
| Enrichment Score (ES) | 0.35764593 |
| Normalized Enrichment Score (NES) | 1.1103059 |
| Nominal p-value | 0.32894737 |
| FDR q-value | 0.8338434 |
| FWER p-Value | 0.957 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
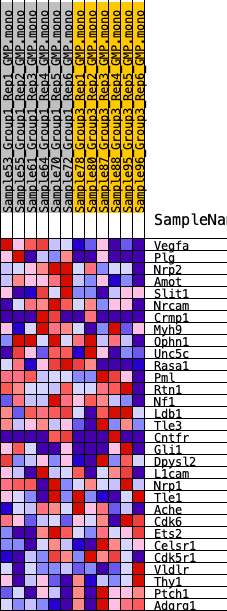
| 1 | Vegfa | 24 | 1.024 | 0.1287 | Yes |
| 2 | Plg | 467 | 0.585 | 0.1841 | Yes |
| 3 | Nrp2 | 969 | 0.484 | 0.2241 | Yes |
| 4 | Amot | 1619 | 0.396 | 0.2468 | Yes |
| 5 | Slit1 | 2326 | 0.343 | 0.2603 | Yes |
| 6 | Nrcam | 2872 | 0.300 | 0.2751 | Yes |
| 7 | Crmp1 | 3017 | 0.292 | 0.3060 | Yes |
| 8 | Myh9 | 3087 | 0.287 | 0.3394 | Yes |
| 9 | Ophn1 | 3451 | 0.265 | 0.3576 | Yes |
| 10 | Unc5c | 4412 | 0.222 | 0.3451 | No |
| 11 | Rasa1 | 5005 | 0.199 | 0.3451 | No |
| 12 | Pml | 5793 | 0.170 | 0.3333 | No |
| 13 | Rtn1 | 6735 | 0.136 | 0.3106 | No |
| 14 | Nf1 | 7033 | 0.126 | 0.3140 | No |
| 15 | Ldb1 | 7283 | 0.118 | 0.3184 | No |
| 16 | Tle3 | 7743 | 0.106 | 0.3124 | No |
| 17 | Cntfr | 8445 | 0.085 | 0.2935 | No |
| 18 | Gli1 | 9280 | 0.060 | 0.2658 | No |
| 19 | Dpysl2 | 10771 | 0.015 | 0.2046 | No |
| 20 | L1cam | 11768 | -0.011 | 0.1637 | No |
| 21 | Nrp1 | 12526 | -0.032 | 0.1357 | No |
| 22 | Tle1 | 13571 | -0.064 | 0.0996 | No |
| 23 | Ache | 14276 | -0.086 | 0.0806 | No |
| 24 | Cdk6 | 15767 | -0.136 | 0.0347 | No |
| 25 | Ets2 | 16378 | -0.158 | 0.0289 | No |
| 26 | Celsr1 | 17612 | -0.201 | 0.0021 | No |
| 27 | Cdk5r1 | 17729 | -0.207 | 0.0234 | No |
| 28 | Vldlr | 19572 | -0.287 | -0.0183 | No |
| 29 | Thy1 | 21056 | -0.372 | -0.0340 | No |
| 30 | Ptch1 | 21463 | -0.399 | -0.0007 | No |
| 31 | Adgrg1 | 23401 | -0.728 | 0.0095 | No |