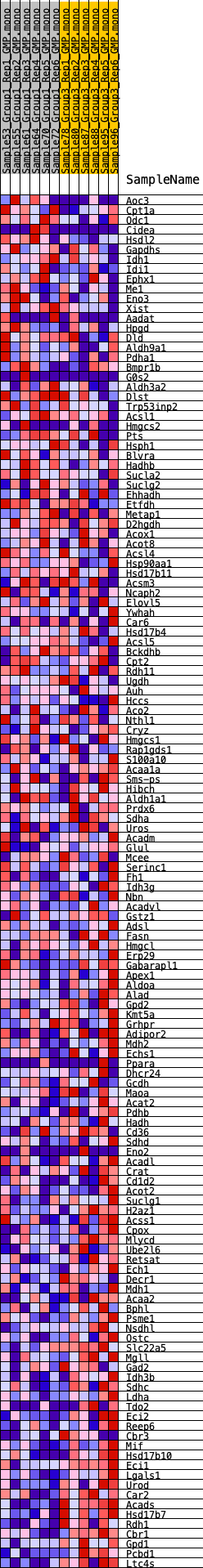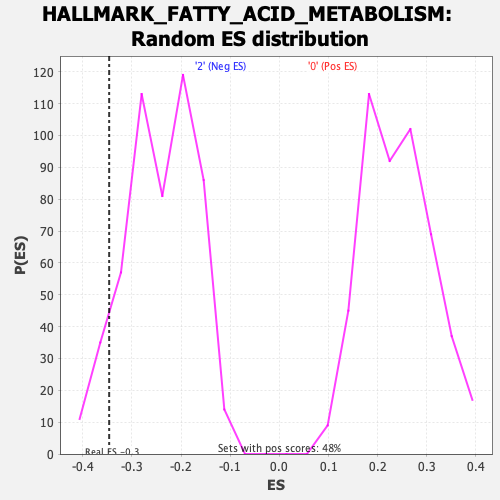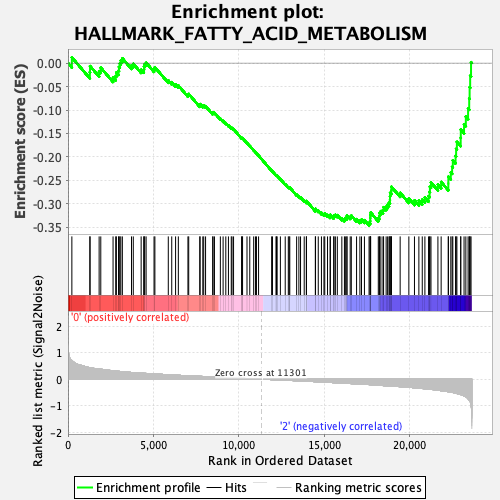
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group3.GMP.mono_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.34639272 |
| Normalized Enrichment Score (NES) | -1.4356302 |
| Nominal p-value | 0.08139535 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.559 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Aoc3 | 226 | 0.704 | 0.0121 | No |
| 2 | Cpt1a | 1279 | 0.432 | -0.0193 | No |
| 3 | Odc1 | 1290 | 0.431 | -0.0064 | No |
| 4 | Cidea | 1821 | 0.379 | -0.0172 | No |
| 5 | Hsdl2 | 1918 | 0.372 | -0.0098 | No |
| 6 | Gapdhs | 2636 | 0.317 | -0.0305 | No |
| 7 | Idh1 | 2792 | 0.306 | -0.0277 | No |
| 8 | Idi1 | 2830 | 0.303 | -0.0199 | No |
| 9 | Ephx1 | 2957 | 0.295 | -0.0161 | No |
| 10 | Me1 | 2982 | 0.294 | -0.0081 | No |
| 11 | Eno3 | 3031 | 0.291 | -0.0011 | No |
| 12 | Xist | 3090 | 0.286 | 0.0053 | No |
| 13 | Aadat | 3180 | 0.280 | 0.0102 | No |
| 14 | Hpgd | 3715 | 0.252 | -0.0048 | No |
| 15 | Dld | 3819 | 0.247 | -0.0015 | No |
| 16 | Aldh9a1 | 4279 | 0.228 | -0.0140 | No |
| 17 | Pdha1 | 4435 | 0.220 | -0.0138 | No |
| 18 | Bmpr1b | 4447 | 0.220 | -0.0074 | No |
| 19 | G0s2 | 4485 | 0.218 | -0.0023 | No |
| 20 | Aldh3a2 | 4569 | 0.216 | 0.0009 | No |
| 21 | Dlst | 5033 | 0.197 | -0.0127 | No |
| 22 | Trp53inp2 | 5083 | 0.195 | -0.0088 | No |
| 23 | Acsl1 | 5867 | 0.167 | -0.0370 | No |
| 24 | Hmgcs2 | 6069 | 0.159 | -0.0406 | No |
| 25 | Pts | 6294 | 0.151 | -0.0455 | No |
| 26 | Hsph1 | 6455 | 0.146 | -0.0478 | No |
| 27 | Blvra | 7017 | 0.127 | -0.0678 | No |
| 28 | Hadhb | 7062 | 0.125 | -0.0658 | No |
| 29 | Sucla2 | 7709 | 0.107 | -0.0900 | No |
| 30 | Suclg2 | 7727 | 0.106 | -0.0874 | No |
| 31 | Ehhadh | 7887 | 0.102 | -0.0910 | No |
| 32 | Etfdh | 7941 | 0.100 | -0.0902 | No |
| 33 | Metap1 | 8048 | 0.096 | -0.0917 | No |
| 34 | D2hgdh | 8463 | 0.085 | -0.1067 | No |
| 35 | Acox1 | 8487 | 0.084 | -0.1051 | No |
| 36 | Acot8 | 8568 | 0.082 | -0.1060 | No |
| 37 | Acsl4 | 8913 | 0.071 | -0.1184 | No |
| 38 | Hsp90aa1 | 9074 | 0.066 | -0.1232 | No |
| 39 | Hsd17b11 | 9232 | 0.062 | -0.1280 | No |
| 40 | Acsm3 | 9386 | 0.057 | -0.1327 | No |
| 41 | Ncaph2 | 9551 | 0.052 | -0.1381 | No |
| 42 | Elovl5 | 9595 | 0.050 | -0.1384 | No |
| 43 | Ywhah | 9685 | 0.047 | -0.1407 | No |
| 44 | Car6 | 10155 | 0.033 | -0.1596 | No |
| 45 | Hsd17b4 | 10181 | 0.033 | -0.1597 | No |
| 46 | Acsl5 | 10212 | 0.032 | -0.1600 | No |
| 47 | Bckdhb | 10472 | 0.024 | -0.1703 | No |
| 48 | Cpt2 | 10634 | 0.019 | -0.1765 | No |
| 49 | Rdh11 | 10863 | 0.013 | -0.1858 | No |
| 50 | Ugdh | 10967 | 0.010 | -0.1899 | No |
| 51 | Auh | 11025 | 0.008 | -0.1921 | No |
| 52 | Hccs | 11149 | 0.005 | -0.1972 | No |
| 53 | Aco2 | 11914 | -0.015 | -0.2293 | No |
| 54 | Nthl1 | 11962 | -0.016 | -0.2308 | No |
| 55 | Cryz | 12168 | -0.022 | -0.2388 | No |
| 56 | Hmgcs1 | 12198 | -0.023 | -0.2393 | No |
| 57 | Rap1gds1 | 12235 | -0.024 | -0.2401 | No |
| 58 | S100a10 | 12425 | -0.030 | -0.2473 | No |
| 59 | Acaa1a | 12708 | -0.037 | -0.2581 | No |
| 60 | Sms-ps | 12888 | -0.043 | -0.2644 | No |
| 61 | Hibch | 12937 | -0.044 | -0.2651 | No |
| 62 | Aldh1a1 | 12975 | -0.045 | -0.2653 | No |
| 63 | Prdx6 | 13375 | -0.058 | -0.2805 | No |
| 64 | Sdha | 13507 | -0.063 | -0.2841 | No |
| 65 | Uros | 13604 | -0.066 | -0.2862 | No |
| 66 | Acadm | 13819 | -0.072 | -0.2930 | No |
| 67 | Glul | 13943 | -0.075 | -0.2960 | No |
| 68 | Mcee | 13952 | -0.076 | -0.2940 | No |
| 69 | Serinc1 | 14469 | -0.092 | -0.3131 | No |
| 70 | Fh1 | 14481 | -0.093 | -0.3107 | No |
| 71 | Idh3g | 14641 | -0.098 | -0.3144 | No |
| 72 | Nbn | 14827 | -0.104 | -0.3191 | No |
| 73 | Acadvl | 14973 | -0.110 | -0.3219 | No |
| 74 | Gstz1 | 15021 | -0.111 | -0.3204 | No |
| 75 | Adsl | 15183 | -0.116 | -0.3237 | No |
| 76 | Fasn | 15336 | -0.121 | -0.3264 | No |
| 77 | Hmgcl | 15348 | -0.121 | -0.3231 | No |
| 78 | Erp29 | 15543 | -0.128 | -0.3274 | No |
| 79 | Gabarapl1 | 15563 | -0.129 | -0.3242 | No |
| 80 | Apex1 | 15642 | -0.132 | -0.3235 | No |
| 81 | Aldoa | 15756 | -0.136 | -0.3241 | No |
| 82 | Alad | 16020 | -0.146 | -0.3308 | No |
| 83 | Gpd2 | 16172 | -0.151 | -0.3325 | No |
| 84 | Kmt5a | 16225 | -0.153 | -0.3300 | No |
| 85 | Grhpr | 16313 | -0.155 | -0.3289 | No |
| 86 | Adipor2 | 16322 | -0.155 | -0.3244 | No |
| 87 | Mdh2 | 16509 | -0.162 | -0.3273 | No |
| 88 | Echs1 | 16574 | -0.165 | -0.3250 | No |
| 89 | Ppara | 16882 | -0.175 | -0.3326 | No |
| 90 | Dhcr24 | 17077 | -0.180 | -0.3353 | No |
| 91 | Gcdh | 17170 | -0.184 | -0.3335 | No |
| 92 | Maoa | 17342 | -0.191 | -0.3349 | No |
| 93 | Acat2 | 17613 | -0.201 | -0.3402 | Yes |
| 94 | Pdhb | 17679 | -0.205 | -0.3366 | Yes |
| 95 | Hadh | 17683 | -0.205 | -0.3304 | Yes |
| 96 | Cd36 | 17686 | -0.205 | -0.3241 | Yes |
| 97 | Sdhd | 17707 | -0.206 | -0.3186 | Yes |
| 98 | Eno2 | 18153 | -0.226 | -0.3305 | Yes |
| 99 | Acadl | 18211 | -0.229 | -0.3259 | Yes |
| 100 | Crat | 18229 | -0.230 | -0.3195 | Yes |
| 101 | Cd1d2 | 18305 | -0.234 | -0.3155 | Yes |
| 102 | Acot2 | 18428 | -0.239 | -0.3133 | Yes |
| 103 | Suclg1 | 18462 | -0.240 | -0.3072 | Yes |
| 104 | H2az1 | 18612 | -0.247 | -0.3060 | Yes |
| 105 | Acss1 | 18699 | -0.251 | -0.3019 | Yes |
| 106 | Cpox | 18780 | -0.254 | -0.2974 | Yes |
| 107 | Mlycd | 18835 | -0.255 | -0.2918 | Yes |
| 108 | Ube2l6 | 18838 | -0.255 | -0.2840 | Yes |
| 109 | Retsat | 18846 | -0.255 | -0.2764 | Yes |
| 110 | Ech1 | 18917 | -0.258 | -0.2714 | Yes |
| 111 | Decr1 | 18920 | -0.258 | -0.2635 | Yes |
| 112 | Mdh1 | 19432 | -0.280 | -0.2766 | Yes |
| 113 | Acaa2 | 19941 | -0.305 | -0.2888 | Yes |
| 114 | Bphl | 20269 | -0.323 | -0.2927 | Yes |
| 115 | Psme1 | 20527 | -0.337 | -0.2932 | Yes |
| 116 | Nsdhl | 20725 | -0.350 | -0.2908 | Yes |
| 117 | Ostc | 20879 | -0.360 | -0.2862 | Yes |
| 118 | Slc22a5 | 21093 | -0.373 | -0.2837 | Yes |
| 119 | Mgll | 21154 | -0.378 | -0.2746 | Yes |
| 120 | Gad2 | 21173 | -0.379 | -0.2636 | Yes |
| 121 | Idh3b | 21235 | -0.382 | -0.2544 | Yes |
| 122 | Sdhc | 21641 | -0.413 | -0.2588 | Yes |
| 123 | Ldha | 21830 | -0.434 | -0.2534 | Yes |
| 124 | Tdo2 | 22250 | -0.471 | -0.2567 | Yes |
| 125 | Eci2 | 22251 | -0.471 | -0.2421 | Yes |
| 126 | Reep6 | 22395 | -0.486 | -0.2331 | Yes |
| 127 | Cbr3 | 22472 | -0.496 | -0.2210 | Yes |
| 128 | Mif | 22513 | -0.501 | -0.2072 | Yes |
| 129 | Hsd17b10 | 22673 | -0.526 | -0.1977 | Yes |
| 130 | Eci1 | 22706 | -0.532 | -0.1826 | Yes |
| 131 | Lgals1 | 22751 | -0.538 | -0.1678 | Yes |
| 132 | Urod | 22973 | -0.578 | -0.1594 | Yes |
| 133 | Car2 | 22979 | -0.579 | -0.1416 | Yes |
| 134 | Acads | 23170 | -0.629 | -0.1303 | Yes |
| 135 | Hsd17b7 | 23275 | -0.665 | -0.1141 | Yes |
| 136 | Rdh1 | 23406 | -0.736 | -0.0969 | Yes |
| 137 | Cbr1 | 23472 | -0.783 | -0.0754 | Yes |
| 138 | Gpd1 | 23496 | -0.804 | -0.0515 | Yes |
| 139 | Pcbd1 | 23528 | -0.844 | -0.0267 | Yes |
| 140 | Ltc4s | 23579 | -0.995 | 0.0019 | Yes |