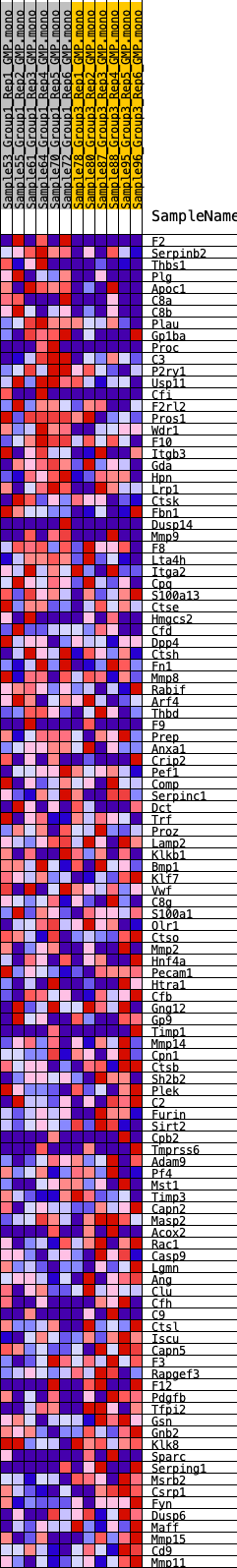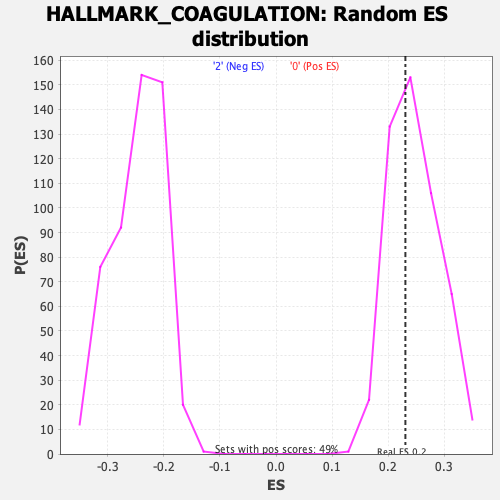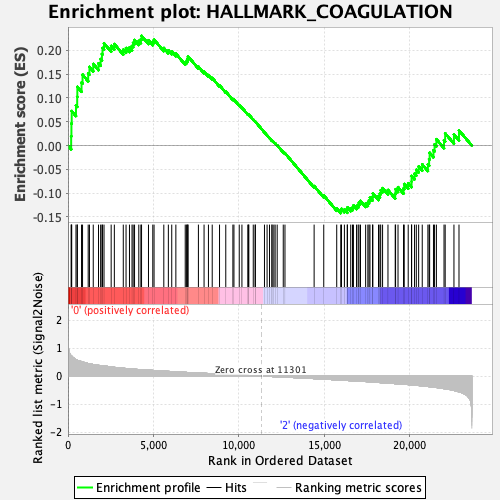
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group3.GMP.mono_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.23051521 |
| Normalized Enrichment Score (NES) | 0.93154114 |
| Nominal p-value | 0.60931176 |
| FDR q-value | 0.9071077 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | F2 | 185 | 0.730 | 0.0198 | Yes |
| 2 | Serpinb2 | 200 | 0.721 | 0.0465 | Yes |
| 3 | Thbs1 | 217 | 0.707 | 0.0726 | Yes |
| 4 | Plg | 467 | 0.585 | 0.0841 | Yes |
| 5 | Apoc1 | 538 | 0.563 | 0.1024 | Yes |
| 6 | C8a | 547 | 0.562 | 0.1234 | Yes |
| 7 | C8b | 792 | 0.515 | 0.1325 | Yes |
| 8 | Plau | 851 | 0.503 | 0.1491 | Yes |
| 9 | Gp1ba | 1181 | 0.445 | 0.1519 | Yes |
| 10 | Proc | 1255 | 0.436 | 0.1653 | Yes |
| 11 | C3 | 1477 | 0.411 | 0.1715 | Yes |
| 12 | P2ry1 | 1785 | 0.382 | 0.1728 | Yes |
| 13 | Usp11 | 1909 | 0.372 | 0.1817 | Yes |
| 14 | Cfi | 1986 | 0.369 | 0.1925 | Yes |
| 15 | F2rl2 | 2020 | 0.367 | 0.2050 | Yes |
| 16 | Pros1 | 2107 | 0.360 | 0.2149 | Yes |
| 17 | Wdr1 | 2525 | 0.323 | 0.2094 | Yes |
| 18 | F10 | 2711 | 0.311 | 0.2133 | Yes |
| 19 | Itgb3 | 3234 | 0.278 | 0.2017 | Yes |
| 20 | Gda | 3396 | 0.268 | 0.2050 | Yes |
| 21 | Hpn | 3596 | 0.258 | 0.2062 | Yes |
| 22 | Lrp1 | 3745 | 0.251 | 0.2094 | Yes |
| 23 | Ctsk | 3811 | 0.248 | 0.2161 | Yes |
| 24 | Fbn1 | 3893 | 0.244 | 0.2218 | Yes |
| 25 | Dusp14 | 4142 | 0.231 | 0.2200 | Yes |
| 26 | Mmp9 | 4264 | 0.229 | 0.2235 | Yes |
| 27 | F8 | 4303 | 0.227 | 0.2305 | Yes |
| 28 | Lta4h | 4708 | 0.210 | 0.2213 | No |
| 29 | Itga2 | 4960 | 0.201 | 0.2182 | No |
| 30 | Cpq | 5034 | 0.197 | 0.2226 | No |
| 31 | S100a13 | 5605 | 0.177 | 0.2050 | No |
| 32 | Ctse | 5871 | 0.166 | 0.2000 | No |
| 33 | Hmgcs2 | 6069 | 0.159 | 0.1977 | No |
| 34 | Cfd | 6310 | 0.150 | 0.1931 | No |
| 35 | Dpp4 | 6858 | 0.132 | 0.1749 | No |
| 36 | Ctsh | 6926 | 0.130 | 0.1769 | No |
| 37 | Fn1 | 6973 | 0.128 | 0.1798 | No |
| 38 | Mmp8 | 7005 | 0.127 | 0.1833 | No |
| 39 | Rabif | 7022 | 0.126 | 0.1874 | No |
| 40 | Arf4 | 7629 | 0.109 | 0.1658 | No |
| 41 | Thbd | 7960 | 0.099 | 0.1555 | No |
| 42 | F9 | 8216 | 0.092 | 0.1481 | No |
| 43 | Prep | 8437 | 0.086 | 0.1420 | No |
| 44 | Anxa1 | 8865 | 0.072 | 0.1266 | No |
| 45 | Crip2 | 9230 | 0.062 | 0.1134 | No |
| 46 | Pef1 | 9646 | 0.049 | 0.0976 | No |
| 47 | Comp | 9708 | 0.047 | 0.0968 | No |
| 48 | Serpinc1 | 10018 | 0.037 | 0.0851 | No |
| 49 | Dct | 10175 | 0.033 | 0.0797 | No |
| 50 | Trf | 10519 | 0.022 | 0.0659 | No |
| 51 | Proz | 10564 | 0.021 | 0.0649 | No |
| 52 | Lamp2 | 10576 | 0.021 | 0.0652 | No |
| 53 | Klkb1 | 10838 | 0.014 | 0.0546 | No |
| 54 | Bmp1 | 10945 | 0.010 | 0.0505 | No |
| 55 | Klf7 | 10963 | 0.010 | 0.0501 | No |
| 56 | Vwf | 11494 | -0.002 | 0.0277 | No |
| 57 | C8g | 11647 | -0.007 | 0.0215 | No |
| 58 | S100a1 | 11789 | -0.011 | 0.0159 | No |
| 59 | Olr1 | 11933 | -0.016 | 0.0104 | No |
| 60 | Ctso | 11934 | -0.016 | 0.0110 | No |
| 61 | Mmp2 | 12042 | -0.019 | 0.0071 | No |
| 62 | Hnf4a | 12117 | -0.021 | 0.0048 | No |
| 63 | Pecam1 | 12238 | -0.024 | 0.0006 | No |
| 64 | Htra1 | 12593 | -0.034 | -0.0132 | No |
| 65 | Cfb | 12693 | -0.037 | -0.0160 | No |
| 66 | Gng12 | 14399 | -0.090 | -0.0851 | No |
| 67 | Gp9 | 14962 | -0.109 | -0.1049 | No |
| 68 | Timp1 | 15721 | -0.135 | -0.1320 | No |
| 69 | Mmp14 | 15957 | -0.144 | -0.1366 | No |
| 70 | Cpn1 | 16001 | -0.145 | -0.1329 | No |
| 71 | Ctsb | 16170 | -0.151 | -0.1344 | No |
| 72 | Sh2b2 | 16337 | -0.156 | -0.1355 | No |
| 73 | Plek | 16347 | -0.156 | -0.1300 | No |
| 74 | C2 | 16540 | -0.164 | -0.1320 | No |
| 75 | Furin | 16644 | -0.167 | -0.1300 | No |
| 76 | Sirt2 | 16698 | -0.169 | -0.1259 | No |
| 77 | Cpb2 | 16881 | -0.175 | -0.1270 | No |
| 78 | Tmprss6 | 16974 | -0.178 | -0.1242 | No |
| 79 | Adam9 | 17023 | -0.179 | -0.1194 | No |
| 80 | Pf4 | 17112 | -0.182 | -0.1163 | No |
| 81 | Mst1 | 17413 | -0.194 | -0.1217 | No |
| 82 | Timp3 | 17543 | -0.199 | -0.1197 | No |
| 83 | Capn2 | 17609 | -0.201 | -0.1148 | No |
| 84 | Masp2 | 17666 | -0.204 | -0.1095 | No |
| 85 | Acox2 | 17814 | -0.211 | -0.1078 | No |
| 86 | Rac1 | 17836 | -0.211 | -0.1007 | No |
| 87 | Casp9 | 18165 | -0.227 | -0.1060 | No |
| 88 | Lgmn | 18248 | -0.231 | -0.1008 | No |
| 89 | Ang | 18295 | -0.233 | -0.0939 | No |
| 90 | Clu | 18404 | -0.238 | -0.0895 | No |
| 91 | Cfh | 18719 | -0.251 | -0.0933 | No |
| 92 | C9 | 19143 | -0.269 | -0.1012 | No |
| 93 | Ctsl | 19164 | -0.270 | -0.0918 | No |
| 94 | Iscu | 19307 | -0.274 | -0.0875 | No |
| 95 | Capn5 | 19628 | -0.290 | -0.0901 | No |
| 96 | F3 | 19670 | -0.292 | -0.0808 | No |
| 97 | Rapgef3 | 19903 | -0.303 | -0.0792 | No |
| 98 | F12 | 20099 | -0.313 | -0.0757 | No |
| 99 | Pdgfb | 20102 | -0.313 | -0.0639 | No |
| 100 | Tfpi2 | 20274 | -0.323 | -0.0590 | No |
| 101 | Gsn | 20377 | -0.328 | -0.0509 | No |
| 102 | Gnb2 | 20514 | -0.336 | -0.0439 | No |
| 103 | Klk8 | 20724 | -0.350 | -0.0396 | No |
| 104 | Sparc | 21055 | -0.372 | -0.0395 | No |
| 105 | Serping1 | 21127 | -0.376 | -0.0283 | No |
| 106 | Msrb2 | 21157 | -0.378 | -0.0152 | No |
| 107 | Csrp1 | 21381 | -0.393 | -0.0099 | No |
| 108 | Fyn | 21445 | -0.397 | 0.0025 | No |
| 109 | Dusp6 | 21552 | -0.406 | 0.0134 | No |
| 110 | Maff | 21998 | -0.447 | 0.0114 | No |
| 111 | Mmp15 | 22066 | -0.453 | 0.0257 | No |
| 112 | Cd9 | 22579 | -0.511 | 0.0232 | No |
| 113 | Mmp11 | 22874 | -0.560 | 0.0319 | No |