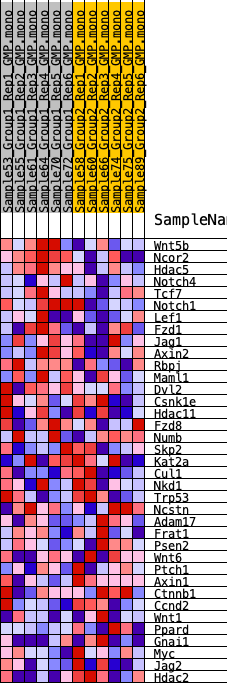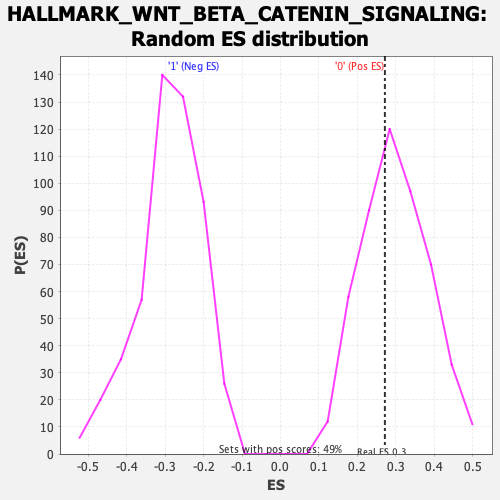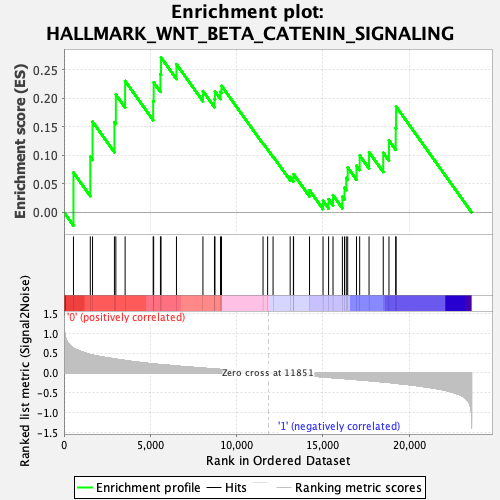
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group2.GMP.mono_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_WNT_BETA_CATENIN_SIGNALING |
| Enrichment Score (ES) | 0.2716319 |
| Normalized Enrichment Score (NES) | 0.9038302 |
| Nominal p-value | 0.63747454 |
| FDR q-value | 0.93012536 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Wnt5b | 548 | 0.620 | 0.0698 | Yes |
| 2 | Ncor2 | 1524 | 0.459 | 0.0975 | Yes |
| 3 | Hdac5 | 1654 | 0.446 | 0.1591 | Yes |
| 4 | Notch4 | 2922 | 0.351 | 0.1580 | Yes |
| 5 | Tcf7 | 3005 | 0.345 | 0.2064 | Yes |
| 6 | Notch1 | 3542 | 0.311 | 0.2304 | Yes |
| 7 | Lef1 | 5169 | 0.225 | 0.1953 | Yes |
| 8 | Fzd1 | 5196 | 0.224 | 0.2278 | Yes |
| 9 | Jag1 | 5589 | 0.207 | 0.2423 | Yes |
| 10 | Axin2 | 5624 | 0.205 | 0.2716 | Yes |
| 11 | Rbpj | 6518 | 0.173 | 0.2597 | No |
| 12 | Maml1 | 8052 | 0.119 | 0.2127 | No |
| 13 | Dvl2 | 8721 | 0.097 | 0.1989 | No |
| 14 | Csnk1e | 8749 | 0.096 | 0.2122 | No |
| 15 | Hdac11 | 9069 | 0.086 | 0.2117 | No |
| 16 | Fzd8 | 9129 | 0.085 | 0.2219 | No |
| 17 | Numb | 11535 | 0.009 | 0.1212 | No |
| 18 | Skp2 | 11801 | 0.002 | 0.1103 | No |
| 19 | Kat2a | 12117 | -0.008 | 0.0980 | No |
| 20 | Cul1 | 13108 | -0.039 | 0.0619 | No |
| 21 | Nkd1 | 13295 | -0.044 | 0.0607 | No |
| 22 | Trp53 | 13306 | -0.045 | 0.0669 | No |
| 23 | Ncstn | 14230 | -0.073 | 0.0387 | No |
| 24 | Adam17 | 15012 | -0.098 | 0.0203 | No |
| 25 | Frat1 | 15335 | -0.109 | 0.0231 | No |
| 26 | Psen2 | 15591 | -0.118 | 0.0301 | No |
| 27 | Wnt6 | 16134 | -0.135 | 0.0274 | No |
| 28 | Ptch1 | 16257 | -0.140 | 0.0432 | No |
| 29 | Axin1 | 16359 | -0.143 | 0.0604 | No |
| 30 | Ctnnb1 | 16442 | -0.145 | 0.0788 | No |
| 31 | Ccnd2 | 16953 | -0.164 | 0.0818 | No |
| 32 | Wnt1 | 17136 | -0.171 | 0.0998 | No |
| 33 | Ppard | 17679 | -0.191 | 0.1055 | No |
| 34 | Gnai1 | 18502 | -0.225 | 0.1045 | No |
| 35 | Myc | 18834 | -0.237 | 0.1261 | No |
| 36 | Jag2 | 19225 | -0.255 | 0.1479 | No |
| 37 | Hdac2 | 19246 | -0.256 | 0.1856 | No |