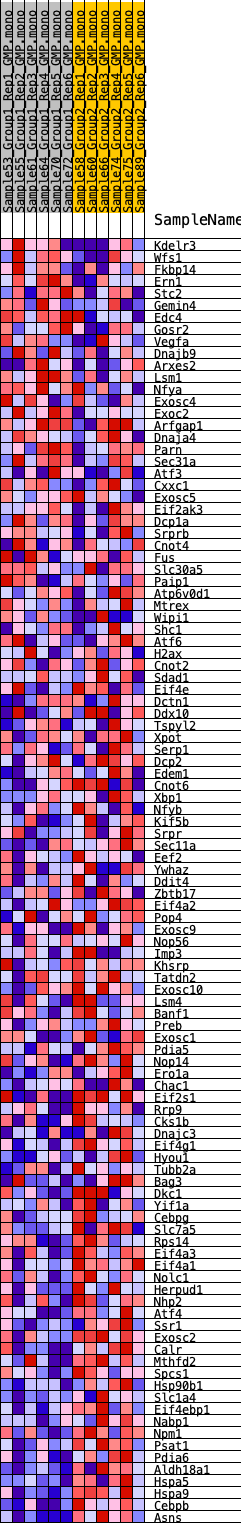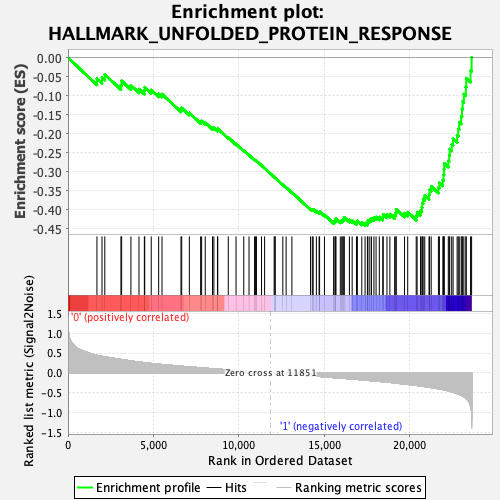
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group2.GMP.mono_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | -0.44230545 |
| Normalized Enrichment Score (NES) | -1.619524 |
| Nominal p-value | 0.053465348 |
| FDR q-value | 0.088305995 |
| FWER p-Value | 0.225 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Kdelr3 | 1693 | 0.442 | -0.0556 | No |
| 2 | Wfs1 | 1989 | 0.418 | -0.0526 | No |
| 3 | Fkbp14 | 2153 | 0.404 | -0.0445 | No |
| 4 | Ern1 | 3097 | 0.339 | -0.0720 | No |
| 5 | Stc2 | 3137 | 0.337 | -0.0612 | No |
| 6 | Gemin4 | 3681 | 0.301 | -0.0731 | No |
| 7 | Edc4 | 4152 | 0.275 | -0.0828 | No |
| 8 | Gosr2 | 4467 | 0.258 | -0.0866 | No |
| 9 | Vegfa | 4489 | 0.258 | -0.0779 | No |
| 10 | Dnajb9 | 4869 | 0.238 | -0.0852 | No |
| 11 | Arxes2 | 5305 | 0.219 | -0.0956 | No |
| 12 | Lsm1 | 5500 | 0.211 | -0.0960 | No |
| 13 | Nfya | 6610 | 0.169 | -0.1369 | No |
| 14 | Exosc4 | 6652 | 0.167 | -0.1324 | No |
| 15 | Exoc2 | 7099 | 0.152 | -0.1457 | No |
| 16 | Arfgap1 | 7759 | 0.130 | -0.1689 | No |
| 17 | Dnaja4 | 7818 | 0.128 | -0.1667 | No |
| 18 | Parn | 8029 | 0.120 | -0.1711 | No |
| 19 | Sec31a | 8453 | 0.105 | -0.1852 | No |
| 20 | Atf3 | 8534 | 0.103 | -0.1848 | No |
| 21 | Cxxc1 | 8745 | 0.096 | -0.1901 | No |
| 22 | Exosc5 | 8762 | 0.096 | -0.1872 | No |
| 23 | Eif2ak3 | 9376 | 0.076 | -0.2105 | No |
| 24 | Dcp1a | 9830 | 0.063 | -0.2274 | No |
| 25 | Srprb | 10282 | 0.048 | -0.2448 | No |
| 26 | Cnot4 | 10590 | 0.038 | -0.2564 | No |
| 27 | Fus | 10914 | 0.029 | -0.2691 | No |
| 28 | Slc30a5 | 10974 | 0.027 | -0.2706 | No |
| 29 | Paip1 | 11025 | 0.025 | -0.2718 | No |
| 30 | Atp6v0d1 | 11323 | 0.015 | -0.2839 | No |
| 31 | Mtrex | 11502 | 0.009 | -0.2911 | No |
| 32 | Wipi1 | 12060 | -0.006 | -0.3146 | No |
| 33 | Shc1 | 12111 | -0.007 | -0.3164 | No |
| 34 | Atf6 | 12116 | -0.007 | -0.3163 | No |
| 35 | H2ax | 12568 | -0.021 | -0.3347 | No |
| 36 | Cnot2 | 12761 | -0.027 | -0.3418 | No |
| 37 | Sdad1 | 13098 | -0.039 | -0.3547 | No |
| 38 | Eif4e | 14190 | -0.072 | -0.3984 | No |
| 39 | Dctn1 | 14292 | -0.075 | -0.3999 | No |
| 40 | Ddx10 | 14341 | -0.076 | -0.3991 | No |
| 41 | Tspyl2 | 14530 | -0.082 | -0.4041 | No |
| 42 | Xpot | 14690 | -0.088 | -0.4076 | No |
| 43 | Serp1 | 14719 | -0.088 | -0.4055 | No |
| 44 | Dcp2 | 15002 | -0.098 | -0.4138 | No |
| 45 | Edem1 | 15538 | -0.117 | -0.4323 | No |
| 46 | Cnot6 | 15613 | -0.120 | -0.4310 | No |
| 47 | Xbp1 | 15665 | -0.121 | -0.4286 | No |
| 48 | Nfyb | 15668 | -0.121 | -0.4242 | No |
| 49 | Kif5b | 15930 | -0.128 | -0.4306 | No |
| 50 | Srpr | 16014 | -0.131 | -0.4292 | No |
| 51 | Sec11a | 16073 | -0.133 | -0.4267 | No |
| 52 | Eef2 | 16133 | -0.135 | -0.4242 | No |
| 53 | Ywhaz | 16163 | -0.136 | -0.4204 | No |
| 54 | Ddit4 | 16463 | -0.146 | -0.4277 | No |
| 55 | Zbtb17 | 16624 | -0.152 | -0.4288 | No |
| 56 | Eif4a2 | 16877 | -0.162 | -0.4335 | No |
| 57 | Pop4 | 16929 | -0.164 | -0.4296 | No |
| 58 | Exosc9 | 17188 | -0.173 | -0.4342 | No |
| 59 | Nop56 | 17380 | -0.180 | -0.4356 | Yes |
| 60 | Imp3 | 17516 | -0.186 | -0.4344 | Yes |
| 61 | Khsrp | 17547 | -0.187 | -0.4288 | Yes |
| 62 | Tatdn2 | 17656 | -0.190 | -0.4263 | Yes |
| 63 | Exosc10 | 17752 | -0.194 | -0.4231 | Yes |
| 64 | Lsm4 | 17898 | -0.200 | -0.4219 | Yes |
| 65 | Banf1 | 18023 | -0.204 | -0.4195 | Yes |
| 66 | Preb | 18212 | -0.212 | -0.4196 | Yes |
| 67 | Exosc1 | 18417 | -0.221 | -0.4201 | Yes |
| 68 | Pdia5 | 18439 | -0.222 | -0.4128 | Yes |
| 69 | Nop14 | 18660 | -0.230 | -0.4136 | Yes |
| 70 | Ero1a | 18831 | -0.237 | -0.4120 | Yes |
| 71 | Chac1 | 19110 | -0.250 | -0.4145 | Yes |
| 72 | Eif2s1 | 19178 | -0.253 | -0.4080 | Yes |
| 73 | Rrp9 | 19203 | -0.255 | -0.3995 | Yes |
| 74 | Cks1b | 19689 | -0.279 | -0.4098 | Yes |
| 75 | Dnajc3 | 19874 | -0.288 | -0.4069 | Yes |
| 76 | Eif4g1 | 20371 | -0.313 | -0.4164 | Yes |
| 77 | Hyou1 | 20423 | -0.316 | -0.4068 | Yes |
| 78 | Tubb2a | 20622 | -0.328 | -0.4030 | Yes |
| 79 | Bag3 | 20705 | -0.334 | -0.3941 | Yes |
| 80 | Dkc1 | 20727 | -0.334 | -0.3826 | Yes |
| 81 | Yif1a | 20778 | -0.337 | -0.3722 | Yes |
| 82 | Cebpg | 20870 | -0.344 | -0.3633 | Yes |
| 83 | Slc7a5 | 21122 | -0.359 | -0.3606 | Yes |
| 84 | Rps14 | 21152 | -0.361 | -0.3484 | Yes |
| 85 | Eif4a3 | 21248 | -0.368 | -0.3388 | Yes |
| 86 | Eif4a1 | 21671 | -0.398 | -0.3420 | Yes |
| 87 | Nolc1 | 21728 | -0.402 | -0.3295 | Yes |
| 88 | Herpud1 | 21930 | -0.419 | -0.3224 | Yes |
| 89 | Nhp2 | 21973 | -0.423 | -0.3085 | Yes |
| 90 | Atf4 | 21995 | -0.426 | -0.2936 | Yes |
| 91 | Ssr1 | 22007 | -0.426 | -0.2782 | Yes |
| 92 | Exosc2 | 22248 | -0.452 | -0.2717 | Yes |
| 93 | Calr | 22302 | -0.457 | -0.2569 | Yes |
| 94 | Mthfd2 | 22332 | -0.461 | -0.2410 | Yes |
| 95 | Spcs1 | 22449 | -0.473 | -0.2284 | Yes |
| 96 | Hsp90b1 | 22529 | -0.486 | -0.2137 | Yes |
| 97 | Slc1a4 | 22764 | -0.519 | -0.2044 | Yes |
| 98 | Eif4ebp1 | 22831 | -0.528 | -0.1876 | Yes |
| 99 | Nabp1 | 22890 | -0.539 | -0.1701 | Yes |
| 100 | Npm1 | 23002 | -0.558 | -0.1541 | Yes |
| 101 | Psat1 | 23051 | -0.570 | -0.1349 | Yes |
| 102 | Pdia6 | 23089 | -0.578 | -0.1150 | Yes |
| 103 | Aldh18a1 | 23160 | -0.595 | -0.0959 | Yes |
| 104 | Hspa5 | 23275 | -0.632 | -0.0773 | Yes |
| 105 | Hspa9 | 23297 | -0.644 | -0.0542 | Yes |
| 106 | Cebpb | 23551 | -0.834 | -0.0340 | Yes |
| 107 | Asns | 23612 | -0.998 | 0.0005 | Yes |