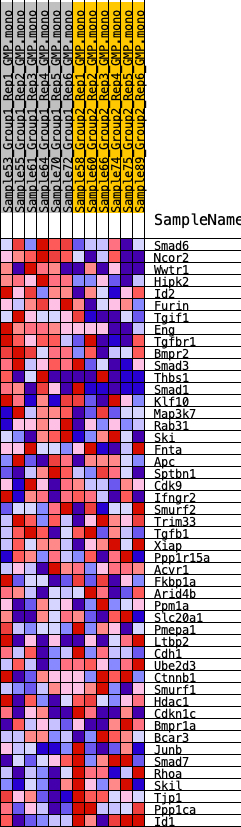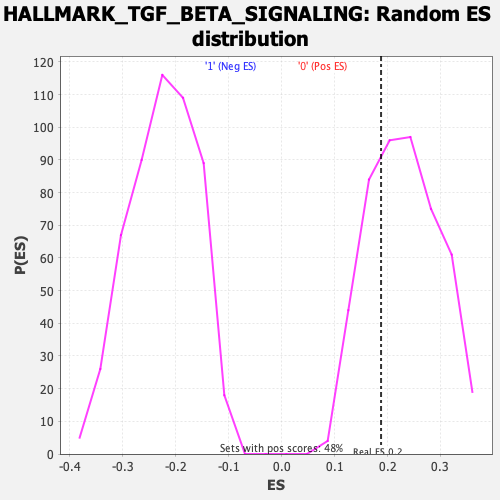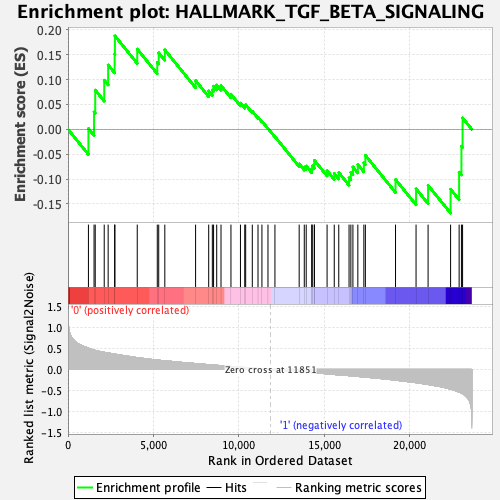
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group2.GMP.mono_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_TGF_BETA_SIGNALING |
| Enrichment Score (ES) | 0.18782349 |
| Normalized Enrichment Score (NES) | 0.8129037 |
| Nominal p-value | 0.72083336 |
| FDR q-value | 0.99737537 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Smad6 | 1195 | 0.505 | 0.0015 | Yes |
| 2 | Ncor2 | 1524 | 0.459 | 0.0350 | Yes |
| 3 | Wwtr1 | 1593 | 0.452 | 0.0789 | Yes |
| 4 | Hipk2 | 2119 | 0.408 | 0.0987 | Yes |
| 5 | Id2 | 2349 | 0.390 | 0.1293 | Yes |
| 6 | Furin | 2729 | 0.364 | 0.1509 | Yes |
| 7 | Tgif1 | 2744 | 0.363 | 0.1878 | Yes |
| 8 | Eng | 4051 | 0.280 | 0.1614 | No |
| 9 | Tgfbr1 | 5223 | 0.223 | 0.1347 | No |
| 10 | Bmpr2 | 5307 | 0.218 | 0.1537 | No |
| 11 | Smad3 | 5664 | 0.204 | 0.1597 | No |
| 12 | Thbs1 | 7464 | 0.140 | 0.0979 | No |
| 13 | Smad1 | 8231 | 0.113 | 0.0771 | No |
| 14 | Klf10 | 8444 | 0.106 | 0.0790 | No |
| 15 | Map3k7 | 8516 | 0.104 | 0.0867 | No |
| 16 | Rab31 | 8700 | 0.098 | 0.0891 | No |
| 17 | Ski | 8951 | 0.090 | 0.0878 | No |
| 18 | Fnta | 9533 | 0.072 | 0.0706 | No |
| 19 | Apc | 10089 | 0.054 | 0.0526 | No |
| 20 | Sptbn1 | 10340 | 0.047 | 0.0469 | No |
| 21 | Cdk9 | 10394 | 0.045 | 0.0493 | No |
| 22 | Ifngr2 | 10782 | 0.033 | 0.0362 | No |
| 23 | Smurf2 | 11116 | 0.022 | 0.0244 | No |
| 24 | Trim33 | 11345 | 0.014 | 0.0162 | No |
| 25 | Tgfb1 | 11699 | 0.005 | 0.0017 | No |
| 26 | Xiap | 12106 | -0.007 | -0.0148 | No |
| 27 | Ppp1r15a | 13524 | -0.052 | -0.0696 | No |
| 28 | Acvr1 | 13823 | -0.061 | -0.0759 | No |
| 29 | Fkbp1a | 13936 | -0.065 | -0.0739 | No |
| 30 | Arid4b | 14256 | -0.074 | -0.0798 | No |
| 31 | Ppm1a | 14291 | -0.075 | -0.0735 | No |
| 32 | Slc20a1 | 14410 | -0.079 | -0.0704 | No |
| 33 | Pmepa1 | 14414 | -0.079 | -0.0624 | No |
| 34 | Ltbp2 | 15157 | -0.103 | -0.0832 | No |
| 35 | Cdh1 | 15576 | -0.118 | -0.0888 | No |
| 36 | Ube2d3 | 15840 | -0.125 | -0.0870 | No |
| 37 | Ctnnb1 | 16442 | -0.145 | -0.0974 | No |
| 38 | Smurf1 | 16548 | -0.149 | -0.0864 | No |
| 39 | Hdac1 | 16668 | -0.153 | -0.0756 | No |
| 40 | Cdkn1c | 16956 | -0.165 | -0.0708 | No |
| 41 | Bmpr1a | 17308 | -0.178 | -0.0673 | No |
| 42 | Bcar3 | 17397 | -0.181 | -0.0524 | No |
| 43 | Junb | 19161 | -0.253 | -0.1011 | No |
| 44 | Smad7 | 20362 | -0.312 | -0.1197 | No |
| 45 | Rhoa | 21066 | -0.355 | -0.1128 | No |
| 46 | Skil | 22383 | -0.465 | -0.1206 | No |
| 47 | Tjp1 | 22882 | -0.537 | -0.0862 | No |
| 48 | Ppp1ca | 23017 | -0.562 | -0.0339 | No |
| 49 | Id1 | 23084 | -0.577 | 0.0229 | No |