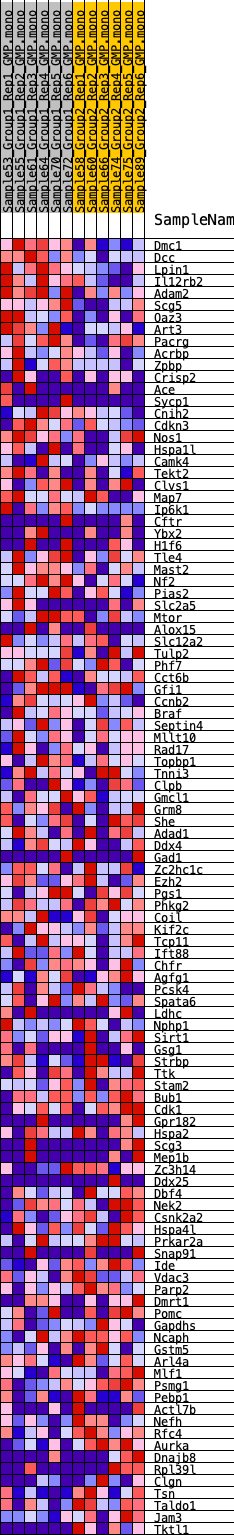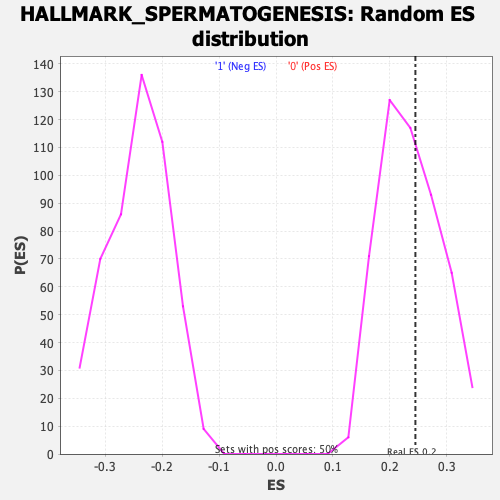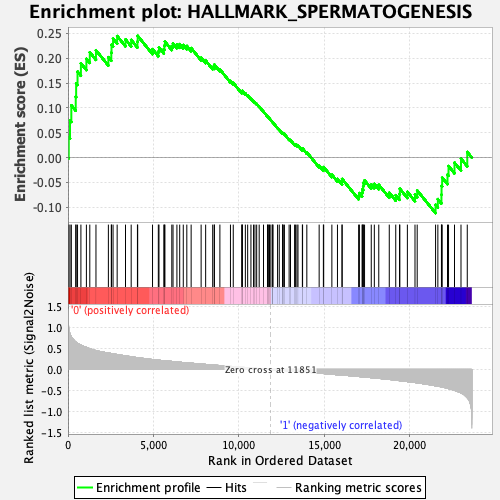
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group2.GMP.mono_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.24502885 |
| Normalized Enrichment Score (NES) | 1.0378414 |
| Nominal p-value | 0.42942345 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.968 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dmc1 | 44 | 0.991 | 0.0412 | Yes |
| 2 | Dcc | 118 | 0.848 | 0.0749 | Yes |
| 3 | Lpin1 | 195 | 0.772 | 0.1052 | Yes |
| 4 | Il12rb2 | 452 | 0.651 | 0.1226 | Yes |
| 5 | Adam2 | 477 | 0.643 | 0.1495 | Yes |
| 6 | Scg5 | 558 | 0.618 | 0.1730 | Yes |
| 7 | Oaz3 | 755 | 0.578 | 0.1898 | Yes |
| 8 | Art3 | 1076 | 0.524 | 0.1989 | Yes |
| 9 | Pacrg | 1275 | 0.493 | 0.2119 | Yes |
| 10 | Acrbp | 1636 | 0.448 | 0.2161 | Yes |
| 11 | Zpbp | 2362 | 0.389 | 0.2021 | Yes |
| 12 | Crisp2 | 2533 | 0.377 | 0.2113 | Yes |
| 13 | Ace | 2546 | 0.376 | 0.2272 | Yes |
| 14 | Sycp1 | 2636 | 0.370 | 0.2395 | Yes |
| 15 | Cnih2 | 2876 | 0.353 | 0.2446 | Yes |
| 16 | Cdkn3 | 3364 | 0.322 | 0.2379 | Yes |
| 17 | Nos1 | 3696 | 0.301 | 0.2369 | Yes |
| 18 | Hspa1l | 4062 | 0.279 | 0.2335 | Yes |
| 19 | Camk4 | 4077 | 0.279 | 0.2450 | Yes |
| 20 | Tekt2 | 4945 | 0.234 | 0.2183 | No |
| 21 | Clvs1 | 5284 | 0.220 | 0.2135 | No |
| 22 | Map7 | 5322 | 0.218 | 0.2214 | No |
| 23 | Ip6k1 | 5606 | 0.206 | 0.2183 | No |
| 24 | Cftr | 5643 | 0.205 | 0.2257 | No |
| 25 | Ybx2 | 5670 | 0.204 | 0.2334 | No |
| 26 | H1f6 | 6073 | 0.190 | 0.2246 | No |
| 27 | Tle4 | 6144 | 0.187 | 0.2297 | No |
| 28 | Mast2 | 6372 | 0.178 | 0.2278 | No |
| 29 | Nf2 | 6535 | 0.172 | 0.2284 | No |
| 30 | Pias2 | 6744 | 0.163 | 0.2267 | No |
| 31 | Slc2a5 | 6951 | 0.157 | 0.2247 | No |
| 32 | Mtor | 7204 | 0.148 | 0.2205 | No |
| 33 | Alox15 | 7788 | 0.129 | 0.2013 | No |
| 34 | Slc12a2 | 8053 | 0.119 | 0.1952 | No |
| 35 | Tulp2 | 8465 | 0.105 | 0.1823 | No |
| 36 | Phf7 | 8559 | 0.102 | 0.1828 | No |
| 37 | Cct6b | 8560 | 0.102 | 0.1872 | No |
| 38 | Gfi1 | 8886 | 0.093 | 0.1774 | No |
| 39 | Ccnb2 | 9504 | 0.073 | 0.1544 | No |
| 40 | Braf | 9668 | 0.068 | 0.1504 | No |
| 41 | Septin4 | 10167 | 0.052 | 0.1314 | No |
| 42 | Mllt10 | 10185 | 0.051 | 0.1329 | No |
| 43 | Rad17 | 10202 | 0.051 | 0.1344 | No |
| 44 | Topbp1 | 10376 | 0.046 | 0.1291 | No |
| 45 | Tnni3 | 10511 | 0.041 | 0.1252 | No |
| 46 | Clpb | 10702 | 0.034 | 0.1186 | No |
| 47 | Gmcl1 | 10855 | 0.031 | 0.1134 | No |
| 48 | Grm8 | 10937 | 0.028 | 0.1112 | No |
| 49 | She | 11038 | 0.024 | 0.1080 | No |
| 50 | Adad1 | 11185 | 0.020 | 0.1027 | No |
| 51 | Ddx4 | 11438 | 0.011 | 0.0924 | No |
| 52 | Gad1 | 11670 | 0.005 | 0.0829 | No |
| 53 | Zc2hc1c | 11700 | 0.005 | 0.0818 | No |
| 54 | Ezh2 | 11716 | 0.004 | 0.0814 | No |
| 55 | Pgs1 | 11788 | 0.002 | 0.0784 | No |
| 56 | Phkg2 | 11826 | 0.001 | 0.0769 | No |
| 57 | Coil | 11940 | -0.002 | 0.0722 | No |
| 58 | Kif2c | 11980 | -0.003 | 0.0707 | No |
| 59 | Tcp11 | 11986 | -0.003 | 0.0706 | No |
| 60 | Ift88 | 12261 | -0.012 | 0.0595 | No |
| 61 | Chfr | 12359 | -0.015 | 0.0560 | No |
| 62 | Agfg1 | 12548 | -0.021 | 0.0489 | No |
| 63 | Pcsk4 | 12554 | -0.021 | 0.0496 | No |
| 64 | Spata6 | 12593 | -0.022 | 0.0490 | No |
| 65 | Ldhc | 12661 | -0.024 | 0.0472 | No |
| 66 | Nphp1 | 12936 | -0.033 | 0.0369 | No |
| 67 | Sirt1 | 13017 | -0.036 | 0.0351 | No |
| 68 | Gsg1 | 13257 | -0.043 | 0.0268 | No |
| 69 | Strbp | 13304 | -0.045 | 0.0268 | No |
| 70 | Ttk | 13392 | -0.047 | 0.0251 | No |
| 71 | Stam2 | 13454 | -0.049 | 0.0247 | No |
| 72 | Bub1 | 13711 | -0.058 | 0.0163 | No |
| 73 | Cdk1 | 13724 | -0.058 | 0.0183 | No |
| 74 | Gpr182 | 13972 | -0.066 | 0.0107 | No |
| 75 | Hspa2 | 14693 | -0.088 | -0.0161 | No |
| 76 | Scg3 | 14940 | -0.095 | -0.0225 | No |
| 77 | Mep1b | 14959 | -0.096 | -0.0191 | No |
| 78 | Zc3h14 | 15437 | -0.113 | -0.0345 | No |
| 79 | Ddx25 | 15764 | -0.124 | -0.0429 | No |
| 80 | Dbf4 | 16032 | -0.132 | -0.0486 | No |
| 81 | Nek2 | 16036 | -0.132 | -0.0430 | No |
| 82 | Csnk2a2 | 17003 | -0.166 | -0.0768 | No |
| 83 | Hspa4l | 17048 | -0.168 | -0.0714 | No |
| 84 | Prkar2a | 17207 | -0.173 | -0.0706 | No |
| 85 | Snap91 | 17225 | -0.174 | -0.0638 | No |
| 86 | Ide | 17270 | -0.176 | -0.0580 | No |
| 87 | Vdac3 | 17277 | -0.176 | -0.0506 | No |
| 88 | Parp2 | 17349 | -0.179 | -0.0458 | No |
| 89 | Dmrt1 | 17741 | -0.194 | -0.0540 | No |
| 90 | Pomc | 17921 | -0.201 | -0.0529 | No |
| 91 | Gapdhs | 18181 | -0.211 | -0.0548 | No |
| 92 | Ncaph | 18795 | -0.236 | -0.0706 | No |
| 93 | Gstm5 | 19174 | -0.253 | -0.0757 | No |
| 94 | Arl4a | 19395 | -0.265 | -0.0735 | No |
| 95 | Mlf1 | 19414 | -0.266 | -0.0627 | No |
| 96 | Psmg1 | 19854 | -0.287 | -0.0689 | No |
| 97 | Pebp1 | 20300 | -0.309 | -0.0744 | No |
| 98 | Actl7b | 20429 | -0.316 | -0.0661 | No |
| 99 | Nefh | 21504 | -0.386 | -0.0951 | No |
| 100 | Rfc4 | 21638 | -0.395 | -0.0836 | No |
| 101 | Aurka | 21847 | -0.412 | -0.0745 | No |
| 102 | Dnajb8 | 21854 | -0.412 | -0.0569 | No |
| 103 | Rpl39l | 21890 | -0.415 | -0.0403 | No |
| 104 | Clgn | 22202 | -0.445 | -0.0342 | No |
| 105 | Tsn | 22262 | -0.453 | -0.0170 | No |
| 106 | Taldo1 | 22611 | -0.497 | -0.0102 | No |
| 107 | Jam3 | 22991 | -0.556 | -0.0022 | No |
| 108 | Tktl1 | 23358 | -0.669 | 0.0113 | No |