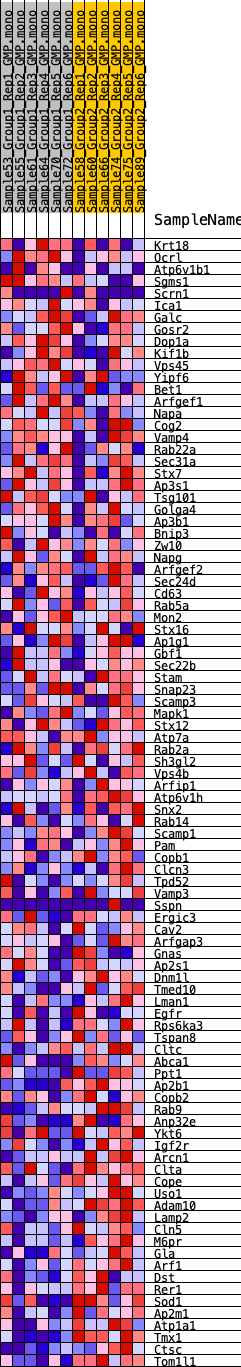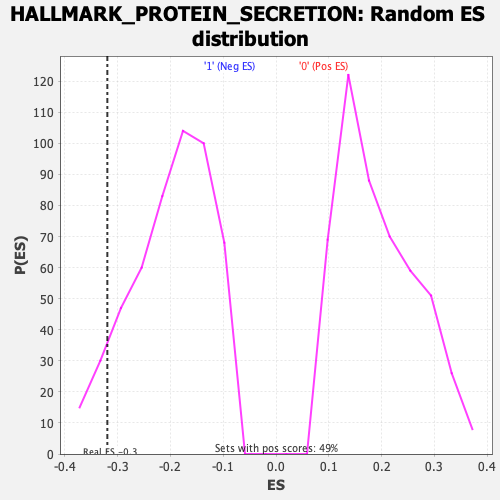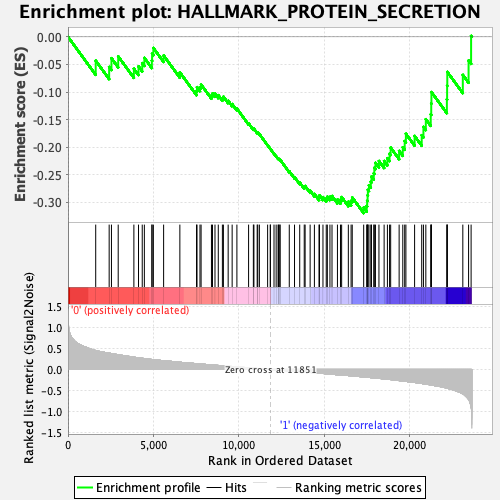
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group2.GMP.mono_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.31933162 |
| Normalized Enrichment Score (NES) | -1.6071736 |
| Nominal p-value | 0.07100592 |
| FDR q-value | 0.085228235 |
| FWER p-Value | 0.244 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Krt18 | 1621 | 0.450 | -0.0432 | No |
| 2 | Ocrl | 2408 | 0.387 | -0.0545 | No |
| 3 | Atp6v1b1 | 2542 | 0.376 | -0.0387 | No |
| 4 | Sgms1 | 2937 | 0.350 | -0.0354 | No |
| 5 | Scrn1 | 3854 | 0.291 | -0.0578 | No |
| 6 | Ica1 | 4125 | 0.276 | -0.0535 | No |
| 7 | Galc | 4343 | 0.265 | -0.0476 | No |
| 8 | Gosr2 | 4467 | 0.258 | -0.0381 | No |
| 9 | Dop1a | 4905 | 0.236 | -0.0431 | No |
| 10 | Kif1b | 4920 | 0.236 | -0.0303 | No |
| 11 | Vps45 | 4994 | 0.232 | -0.0201 | No |
| 12 | Yipf6 | 5594 | 0.206 | -0.0338 | No |
| 13 | Bet1 | 6544 | 0.171 | -0.0644 | No |
| 14 | Arfgef1 | 7519 | 0.138 | -0.0978 | No |
| 15 | Napa | 7547 | 0.137 | -0.0912 | No |
| 16 | Cog2 | 7721 | 0.131 | -0.0910 | No |
| 17 | Vamp4 | 7786 | 0.129 | -0.0864 | No |
| 18 | Rab22a | 8392 | 0.108 | -0.1060 | No |
| 19 | Sec31a | 8453 | 0.105 | -0.1025 | No |
| 20 | Stx7 | 8599 | 0.101 | -0.1029 | No |
| 21 | Ap3s1 | 8792 | 0.095 | -0.1056 | No |
| 22 | Tsg101 | 9032 | 0.088 | -0.1108 | No |
| 23 | Golga4 | 9091 | 0.086 | -0.1084 | No |
| 24 | Ap3b1 | 9370 | 0.077 | -0.1158 | No |
| 25 | Bnip3 | 9603 | 0.070 | -0.1217 | No |
| 26 | Zw10 | 9881 | 0.061 | -0.1300 | No |
| 27 | Napg | 10565 | 0.039 | -0.1568 | No |
| 28 | Arfgef2 | 10844 | 0.031 | -0.1668 | No |
| 29 | Sec24d | 10876 | 0.030 | -0.1664 | No |
| 30 | Cd63 | 11068 | 0.023 | -0.1732 | No |
| 31 | Rab5a | 11102 | 0.023 | -0.1733 | No |
| 32 | Mon2 | 11196 | 0.019 | -0.1762 | No |
| 33 | Stx16 | 11678 | 0.005 | -0.1963 | No |
| 34 | Ap1g1 | 11833 | 0.001 | -0.2028 | No |
| 35 | Gbf1 | 11842 | 0.000 | -0.2031 | No |
| 36 | Sec22b | 12052 | -0.006 | -0.2117 | No |
| 37 | Stam | 12177 | -0.010 | -0.2164 | No |
| 38 | Snap23 | 12290 | -0.013 | -0.2204 | No |
| 39 | Scamp3 | 12317 | -0.014 | -0.2207 | No |
| 40 | Mapk1 | 12389 | -0.016 | -0.2228 | No |
| 41 | Stx12 | 12409 | -0.017 | -0.2226 | No |
| 42 | Atp7a | 12945 | -0.033 | -0.2435 | No |
| 43 | Rab2a | 13252 | -0.043 | -0.2540 | No |
| 44 | Sh3gl2 | 13556 | -0.053 | -0.2639 | No |
| 45 | Vps4b | 13820 | -0.061 | -0.2716 | No |
| 46 | Arfip1 | 13872 | -0.063 | -0.2701 | No |
| 47 | Atp6v1h | 14165 | -0.071 | -0.2785 | No |
| 48 | Snx2 | 14415 | -0.079 | -0.2846 | No |
| 49 | Rab14 | 14685 | -0.087 | -0.2910 | No |
| 50 | Scamp1 | 14712 | -0.088 | -0.2871 | No |
| 51 | Pam | 14916 | -0.094 | -0.2903 | No |
| 52 | Copb1 | 15108 | -0.102 | -0.2927 | No |
| 53 | Clcn3 | 15172 | -0.104 | -0.2894 | No |
| 54 | Tpd52 | 15324 | -0.109 | -0.2896 | No |
| 55 | Vamp3 | 15445 | -0.113 | -0.2883 | No |
| 56 | Sspn | 15763 | -0.124 | -0.2947 | No |
| 57 | Ergic3 | 15941 | -0.128 | -0.2948 | No |
| 58 | Cav2 | 16009 | -0.131 | -0.2902 | No |
| 59 | Arfgap3 | 16400 | -0.144 | -0.2986 | No |
| 60 | Gnas | 16562 | -0.150 | -0.2968 | No |
| 61 | Ap2s1 | 16629 | -0.152 | -0.2909 | No |
| 62 | Dnm1l | 17298 | -0.177 | -0.3092 | Yes |
| 63 | Tmed10 | 17477 | -0.185 | -0.3062 | Yes |
| 64 | Lman1 | 17503 | -0.185 | -0.2967 | Yes |
| 65 | Egfr | 17528 | -0.186 | -0.2871 | Yes |
| 66 | Rps6ka3 | 17544 | -0.187 | -0.2771 | Yes |
| 67 | Tspan8 | 17606 | -0.189 | -0.2689 | Yes |
| 68 | Cltc | 17715 | -0.193 | -0.2625 | Yes |
| 69 | Abca1 | 17754 | -0.194 | -0.2530 | Yes |
| 70 | Ppt1 | 17888 | -0.200 | -0.2472 | Yes |
| 71 | Ap2b1 | 17920 | -0.201 | -0.2371 | Yes |
| 72 | Copb2 | 17986 | -0.203 | -0.2282 | Yes |
| 73 | Rab9 | 18191 | -0.211 | -0.2249 | Yes |
| 74 | Anp32e | 18494 | -0.225 | -0.2249 | Yes |
| 75 | Ykt6 | 18686 | -0.231 | -0.2198 | Yes |
| 76 | Igf2r | 18812 | -0.236 | -0.2116 | Yes |
| 77 | Arcn1 | 18877 | -0.240 | -0.2007 | Yes |
| 78 | Clta | 19372 | -0.263 | -0.2066 | Yes |
| 79 | Cope | 19586 | -0.275 | -0.2000 | Yes |
| 80 | Uso1 | 19698 | -0.280 | -0.1887 | Yes |
| 81 | Adam10 | 19768 | -0.282 | -0.1755 | Yes |
| 82 | Lamp2 | 20277 | -0.308 | -0.1795 | Yes |
| 83 | Cln5 | 20693 | -0.333 | -0.1782 | Yes |
| 84 | M6pr | 20794 | -0.338 | -0.1631 | Yes |
| 85 | Gla | 20934 | -0.348 | -0.1492 | Yes |
| 86 | Arf1 | 21221 | -0.366 | -0.1404 | Yes |
| 87 | Dst | 21250 | -0.368 | -0.1206 | Yes |
| 88 | Rer1 | 21262 | -0.369 | -0.1000 | Yes |
| 89 | Sod1 | 22165 | -0.441 | -0.1132 | Yes |
| 90 | Ap2m1 | 22180 | -0.443 | -0.0885 | Yes |
| 91 | Atp1a1 | 22190 | -0.444 | -0.0635 | Yes |
| 92 | Tmx1 | 23098 | -0.580 | -0.0689 | Yes |
| 93 | Ctsc | 23435 | -0.707 | -0.0428 | Yes |
| 94 | Tom1l1 | 23583 | -0.891 | 0.0017 | Yes |