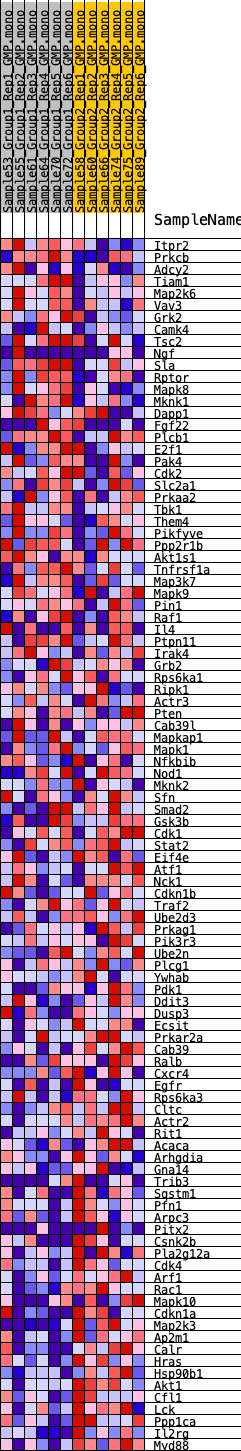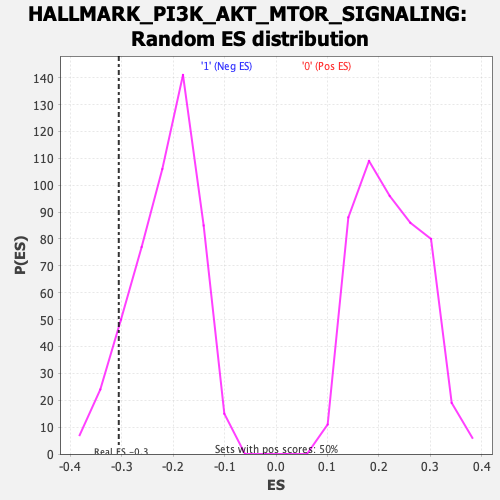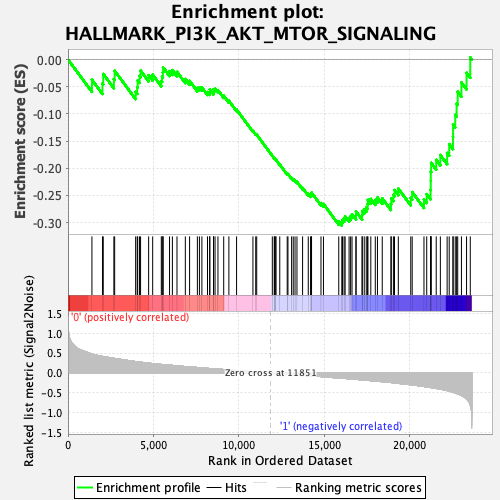
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group2.GMP.mono_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.30542767 |
| Normalized Enrichment Score (NES) | -1.4232999 |
| Nominal p-value | 0.08118812 |
| FDR q-value | 0.12990054 |
| FWER p-Value | 0.5 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Itpr2 | 1400 | 0.477 | -0.0373 | No |
| 2 | Prkcb | 2017 | 0.415 | -0.0441 | No |
| 3 | Adcy2 | 2062 | 0.412 | -0.0267 | No |
| 4 | Tiam1 | 2684 | 0.367 | -0.0359 | No |
| 5 | Map2k6 | 2730 | 0.364 | -0.0208 | No |
| 6 | Vav3 | 3957 | 0.286 | -0.0596 | No |
| 7 | Grk2 | 4049 | 0.280 | -0.0504 | No |
| 8 | Camk4 | 4077 | 0.279 | -0.0385 | No |
| 9 | Tsc2 | 4185 | 0.272 | -0.0304 | No |
| 10 | Ngf | 4245 | 0.269 | -0.0203 | No |
| 11 | Sla | 4717 | 0.246 | -0.0289 | No |
| 12 | Rptor | 4954 | 0.234 | -0.0280 | No |
| 13 | Mapk8 | 5458 | 0.213 | -0.0394 | No |
| 14 | Mknk1 | 5504 | 0.211 | -0.0315 | No |
| 15 | Dapp1 | 5549 | 0.209 | -0.0236 | No |
| 16 | Fgf22 | 5565 | 0.208 | -0.0145 | No |
| 17 | Plcb1 | 5940 | 0.195 | -0.0214 | No |
| 18 | E2f1 | 6107 | 0.189 | -0.0196 | No |
| 19 | Pak4 | 6376 | 0.178 | -0.0227 | No |
| 20 | Cdk2 | 6864 | 0.160 | -0.0359 | No |
| 21 | Slc2a1 | 7113 | 0.151 | -0.0394 | No |
| 22 | Prkaa2 | 7569 | 0.136 | -0.0523 | No |
| 23 | Tbk1 | 7688 | 0.133 | -0.0512 | No |
| 24 | Them4 | 7833 | 0.127 | -0.0513 | No |
| 25 | Pikfyve | 8161 | 0.115 | -0.0599 | No |
| 26 | Ppp2r1b | 8288 | 0.111 | -0.0600 | No |
| 27 | Akt1s1 | 8297 | 0.111 | -0.0552 | No |
| 28 | Tnfrsf1a | 8513 | 0.104 | -0.0595 | No |
| 29 | Map3k7 | 8516 | 0.104 | -0.0548 | No |
| 30 | Mapk9 | 8603 | 0.101 | -0.0537 | No |
| 31 | Pin1 | 8772 | 0.096 | -0.0564 | No |
| 32 | Raf1 | 9109 | 0.085 | -0.0667 | No |
| 33 | Il4 | 9412 | 0.075 | -0.0760 | No |
| 34 | Ptpn11 | 9862 | 0.062 | -0.0922 | No |
| 35 | Irak4 | 10819 | 0.032 | -0.1314 | No |
| 36 | Grb2 | 10982 | 0.026 | -0.1370 | No |
| 37 | Rps6ka1 | 11040 | 0.024 | -0.1383 | No |
| 38 | Ripk1 | 11953 | -0.002 | -0.1770 | No |
| 39 | Actr3 | 12069 | -0.006 | -0.1816 | No |
| 40 | Pten | 12075 | -0.006 | -0.1815 | No |
| 41 | Cab39l | 12151 | -0.009 | -0.1843 | No |
| 42 | Mapkap1 | 12165 | -0.009 | -0.1844 | No |
| 43 | Mapk1 | 12389 | -0.016 | -0.1931 | No |
| 44 | Nfkbib | 12824 | -0.029 | -0.2102 | No |
| 45 | Nod1 | 12868 | -0.031 | -0.2106 | No |
| 46 | Mknk2 | 13081 | -0.038 | -0.2179 | No |
| 47 | Sfn | 13177 | -0.041 | -0.2200 | No |
| 48 | Smad2 | 13282 | -0.044 | -0.2224 | No |
| 49 | Gsk3b | 13395 | -0.047 | -0.2249 | No |
| 50 | Cdk1 | 13724 | -0.058 | -0.2361 | No |
| 51 | Stat2 | 14056 | -0.068 | -0.2470 | No |
| 52 | Eif4e | 14190 | -0.072 | -0.2493 | No |
| 53 | Atf1 | 14242 | -0.073 | -0.2481 | No |
| 54 | Nck1 | 14248 | -0.074 | -0.2448 | No |
| 55 | Cdkn1b | 14800 | -0.091 | -0.2640 | No |
| 56 | Traf2 | 14945 | -0.095 | -0.2657 | No |
| 57 | Ube2d3 | 15840 | -0.125 | -0.2979 | No |
| 58 | Prkag1 | 16019 | -0.131 | -0.2993 | Yes |
| 59 | Pik3r3 | 16061 | -0.133 | -0.2948 | Yes |
| 60 | Ube2n | 16152 | -0.136 | -0.2923 | Yes |
| 61 | Plcg1 | 16212 | -0.138 | -0.2884 | Yes |
| 62 | Ywhab | 16444 | -0.146 | -0.2914 | Yes |
| 63 | Pdk1 | 16534 | -0.149 | -0.2882 | Yes |
| 64 | Ddit3 | 16616 | -0.152 | -0.2846 | Yes |
| 65 | Dusp3 | 16848 | -0.160 | -0.2869 | Yes |
| 66 | Ecsit | 16850 | -0.160 | -0.2794 | Yes |
| 67 | Prkar2a | 17207 | -0.173 | -0.2865 | Yes |
| 68 | Cab39 | 17212 | -0.174 | -0.2786 | Yes |
| 69 | Ralb | 17331 | -0.178 | -0.2752 | Yes |
| 70 | Cxcr4 | 17449 | -0.184 | -0.2717 | Yes |
| 71 | Egfr | 17528 | -0.186 | -0.2663 | Yes |
| 72 | Rps6ka3 | 17544 | -0.187 | -0.2582 | Yes |
| 73 | Cltc | 17715 | -0.193 | -0.2564 | Yes |
| 74 | Actr2 | 17977 | -0.203 | -0.2580 | Yes |
| 75 | Rit1 | 18104 | -0.208 | -0.2537 | Yes |
| 76 | Acaca | 18384 | -0.219 | -0.2553 | Yes |
| 77 | Arhgdia | 18891 | -0.240 | -0.2656 | Yes |
| 78 | Gna14 | 18913 | -0.241 | -0.2553 | Yes |
| 79 | Trib3 | 19037 | -0.247 | -0.2490 | Yes |
| 80 | Sqstm1 | 19101 | -0.249 | -0.2400 | Yes |
| 81 | Pfn1 | 19324 | -0.260 | -0.2373 | Yes |
| 82 | Arpc3 | 20052 | -0.297 | -0.2544 | Yes |
| 83 | Pitx2 | 20136 | -0.302 | -0.2438 | Yes |
| 84 | Csnk2b | 20825 | -0.340 | -0.2571 | Yes |
| 85 | Pla2g12a | 20986 | -0.351 | -0.2476 | Yes |
| 86 | Cdk4 | 21212 | -0.366 | -0.2401 | Yes |
| 87 | Arf1 | 21221 | -0.366 | -0.2233 | Yes |
| 88 | Rac1 | 21223 | -0.366 | -0.2063 | Yes |
| 89 | Mapk10 | 21246 | -0.368 | -0.1900 | Yes |
| 90 | Cdkn1a | 21542 | -0.388 | -0.1845 | Yes |
| 91 | Map2k3 | 21784 | -0.407 | -0.1757 | Yes |
| 92 | Ap2m1 | 22180 | -0.443 | -0.1718 | Yes |
| 93 | Calr | 22302 | -0.457 | -0.1556 | Yes |
| 94 | Hras | 22522 | -0.485 | -0.1422 | Yes |
| 95 | Hsp90b1 | 22529 | -0.486 | -0.1198 | Yes |
| 96 | Akt1 | 22654 | -0.502 | -0.1017 | Yes |
| 97 | Cfl1 | 22735 | -0.515 | -0.0810 | Yes |
| 98 | Lck | 22799 | -0.524 | -0.0592 | Yes |
| 99 | Ppp1ca | 23017 | -0.562 | -0.0423 | Yes |
| 100 | Il2rg | 23317 | -0.653 | -0.0245 | Yes |
| 101 | Myd88 | 23534 | -0.803 | 0.0038 | Yes |