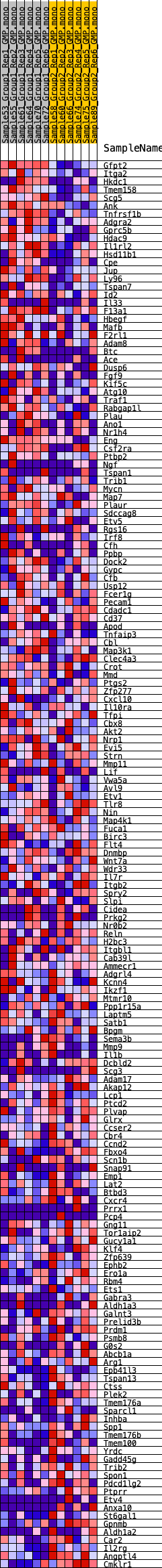
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group2.GMP.mono_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
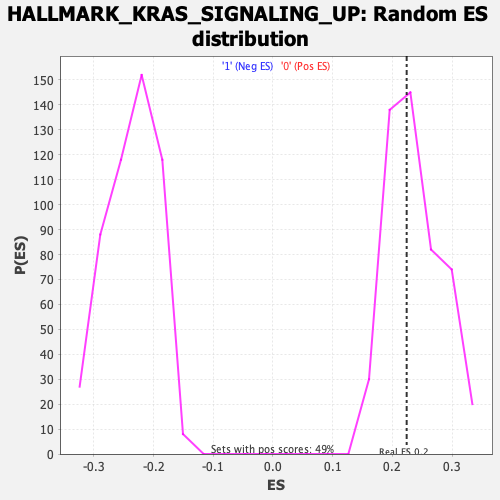
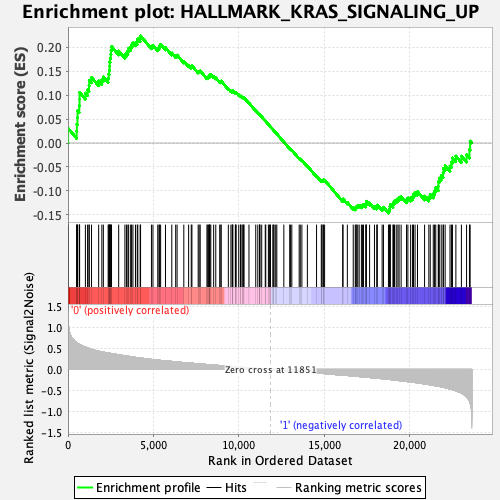
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | 0.2238536 |
| Normalized Enrichment Score (NES) | 0.9491992 |
| Nominal p-value | 0.5337423 |
| FDR q-value | 0.97103274 |
| FWER p-Value | 0.989 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gfpt2 | 6 | 1.285 | 0.0308 | Yes |
| 2 | Itga2 | 505 | 0.633 | 0.0248 | Yes |
| 3 | Hkdc1 | 518 | 0.629 | 0.0395 | Yes |
| 4 | Tmem158 | 552 | 0.619 | 0.0530 | Yes |
| 5 | Scg5 | 558 | 0.618 | 0.0677 | Yes |
| 6 | Ank | 658 | 0.597 | 0.0779 | Yes |
| 7 | Tnfrsf1b | 675 | 0.594 | 0.0916 | Yes |
| 8 | Adgra2 | 678 | 0.594 | 0.1058 | Yes |
| 9 | Gprc5b | 1017 | 0.533 | 0.1043 | Yes |
| 10 | Hdac9 | 1145 | 0.513 | 0.1113 | Yes |
| 11 | Il1rl2 | 1229 | 0.499 | 0.1198 | Yes |
| 12 | Hsd11b1 | 1241 | 0.498 | 0.1313 | Yes |
| 13 | Cpe | 1376 | 0.479 | 0.1372 | Yes |
| 14 | Jup | 1794 | 0.432 | 0.1298 | Yes |
| 15 | Ly96 | 1980 | 0.418 | 0.1321 | Yes |
| 16 | Tspan7 | 2068 | 0.412 | 0.1383 | Yes |
| 17 | Id2 | 2349 | 0.390 | 0.1358 | Yes |
| 18 | Il33 | 2380 | 0.388 | 0.1439 | Yes |
| 19 | F13a1 | 2412 | 0.387 | 0.1519 | Yes |
| 20 | Hbegf | 2435 | 0.386 | 0.1603 | Yes |
| 21 | Mafb | 2439 | 0.385 | 0.1694 | Yes |
| 22 | F2rl1 | 2467 | 0.382 | 0.1775 | Yes |
| 23 | Adam8 | 2502 | 0.380 | 0.1852 | Yes |
| 24 | Btc | 2517 | 0.378 | 0.1938 | Yes |
| 25 | Ace | 2546 | 0.376 | 0.2017 | Yes |
| 26 | Dusp6 | 2965 | 0.348 | 0.1922 | Yes |
| 27 | Fgf9 | 3329 | 0.325 | 0.1846 | Yes |
| 28 | Kif5c | 3420 | 0.320 | 0.1885 | Yes |
| 29 | Atg10 | 3504 | 0.313 | 0.1925 | Yes |
| 30 | Traf1 | 3549 | 0.311 | 0.1981 | Yes |
| 31 | Rabgap1l | 3666 | 0.303 | 0.2005 | Yes |
| 32 | Plau | 3720 | 0.300 | 0.2055 | Yes |
| 33 | Ano1 | 3799 | 0.295 | 0.2093 | Yes |
| 34 | Nr1h4 | 3948 | 0.286 | 0.2098 | Yes |
| 35 | Eng | 4051 | 0.280 | 0.2123 | Yes |
| 36 | Csf2ra | 4083 | 0.278 | 0.2177 | Yes |
| 37 | Ptbp2 | 4222 | 0.270 | 0.2183 | Yes |
| 38 | Ngf | 4245 | 0.269 | 0.2239 | Yes |
| 39 | Tspan1 | 4882 | 0.238 | 0.2025 | No |
| 40 | Trib1 | 4970 | 0.234 | 0.2044 | No |
| 41 | Mycn | 5251 | 0.221 | 0.1978 | No |
| 42 | Map7 | 5322 | 0.218 | 0.2001 | No |
| 43 | Plaur | 5359 | 0.217 | 0.2038 | No |
| 44 | Sdccag8 | 5419 | 0.214 | 0.2064 | No |
| 45 | Etv5 | 5703 | 0.204 | 0.1993 | No |
| 46 | Rgs16 | 6072 | 0.190 | 0.1882 | No |
| 47 | Irf8 | 6300 | 0.181 | 0.1829 | No |
| 48 | Cfh | 6382 | 0.178 | 0.1837 | No |
| 49 | Ppbp | 6775 | 0.162 | 0.1709 | No |
| 50 | Dock2 | 7056 | 0.153 | 0.1627 | No |
| 51 | Gypc | 7210 | 0.148 | 0.1598 | No |
| 52 | Cfb | 7250 | 0.147 | 0.1616 | No |
| 53 | Usp12 | 7615 | 0.135 | 0.1494 | No |
| 54 | Fcer1g | 7672 | 0.133 | 0.1502 | No |
| 55 | Pecam1 | 7734 | 0.131 | 0.1508 | No |
| 56 | Cdadc1 | 8128 | 0.117 | 0.1368 | No |
| 57 | Cd37 | 8178 | 0.115 | 0.1375 | No |
| 58 | Apod | 8213 | 0.113 | 0.1388 | No |
| 59 | Tnfaip3 | 8269 | 0.112 | 0.1391 | No |
| 60 | Cbl | 8279 | 0.112 | 0.1414 | No |
| 61 | Map3k1 | 8302 | 0.111 | 0.1432 | No |
| 62 | Clec4a3 | 8354 | 0.109 | 0.1436 | No |
| 63 | Crot | 8521 | 0.103 | 0.1390 | No |
| 64 | Mmd | 8645 | 0.099 | 0.1362 | No |
| 65 | Ptgs2 | 8875 | 0.093 | 0.1287 | No |
| 66 | Zfp277 | 8948 | 0.091 | 0.1278 | No |
| 67 | Cxcl10 | 8956 | 0.090 | 0.1297 | No |
| 68 | Il10ra | 9380 | 0.076 | 0.1135 | No |
| 69 | Tfpi | 9532 | 0.072 | 0.1088 | No |
| 70 | Cbx8 | 9609 | 0.069 | 0.1072 | No |
| 71 | Akt2 | 9620 | 0.069 | 0.1084 | No |
| 72 | Nrp1 | 9646 | 0.068 | 0.1090 | No |
| 73 | Evi5 | 9789 | 0.064 | 0.1045 | No |
| 74 | Strn | 9805 | 0.064 | 0.1054 | No |
| 75 | Mmp11 | 9837 | 0.062 | 0.1056 | No |
| 76 | Lif | 9987 | 0.057 | 0.1006 | No |
| 77 | Vwa5a | 10087 | 0.054 | 0.0977 | No |
| 78 | Avl9 | 10107 | 0.054 | 0.0982 | No |
| 79 | Etv1 | 10183 | 0.051 | 0.0962 | No |
| 80 | Tlr8 | 10236 | 0.050 | 0.0952 | No |
| 81 | Nin | 10276 | 0.048 | 0.0947 | No |
| 82 | Map4k1 | 10296 | 0.048 | 0.0951 | No |
| 83 | Fuca1 | 10587 | 0.038 | 0.0836 | No |
| 84 | Birc3 | 10983 | 0.026 | 0.0674 | No |
| 85 | Flt4 | 11104 | 0.023 | 0.0628 | No |
| 86 | Dnmbp | 11209 | 0.019 | 0.0589 | No |
| 87 | Wnt7a | 11215 | 0.019 | 0.0591 | No |
| 88 | Wdr33 | 11226 | 0.018 | 0.0591 | No |
| 89 | Il7r | 11332 | 0.015 | 0.0550 | No |
| 90 | Itgb2 | 11540 | 0.008 | 0.0464 | No |
| 91 | Spry2 | 11569 | 0.008 | 0.0454 | No |
| 92 | Slpi | 11724 | 0.004 | 0.0389 | No |
| 93 | Cidea | 11780 | 0.002 | 0.0366 | No |
| 94 | Prkg2 | 11819 | 0.001 | 0.0350 | No |
| 95 | Nr0b2 | 11847 | 0.000 | 0.0339 | No |
| 96 | Reln | 11982 | -0.003 | 0.0282 | No |
| 97 | H2bc3 | 12041 | -0.006 | 0.0259 | No |
| 98 | Itgbl1 | 12046 | -0.006 | 0.0259 | No |
| 99 | Cab39l | 12151 | -0.009 | 0.0216 | No |
| 100 | Ammecr1 | 12216 | -0.011 | 0.0192 | No |
| 101 | Adgrl4 | 12627 | -0.023 | 0.0022 | No |
| 102 | Kcnn4 | 12970 | -0.034 | -0.0115 | No |
| 103 | Ikzf1 | 13023 | -0.036 | -0.0129 | No |
| 104 | Mtmr10 | 13094 | -0.038 | -0.0149 | No |
| 105 | Ppp1r15a | 13524 | -0.052 | -0.0320 | No |
| 106 | Laptm5 | 13589 | -0.054 | -0.0334 | No |
| 107 | Satb1 | 13683 | -0.057 | -0.0360 | No |
| 108 | Bpgm | 14004 | -0.067 | -0.0480 | No |
| 109 | Sema3b | 14541 | -0.083 | -0.0689 | No |
| 110 | Mmp9 | 14818 | -0.091 | -0.0785 | No |
| 111 | Il1b | 14854 | -0.092 | -0.0777 | No |
| 112 | Dcbld2 | 14924 | -0.094 | -0.0784 | No |
| 113 | Scg3 | 14940 | -0.095 | -0.0767 | No |
| 114 | Adam17 | 15012 | -0.098 | -0.0774 | No |
| 115 | Akap12 | 16070 | -0.133 | -0.1193 | No |
| 116 | Lcp1 | 16089 | -0.134 | -0.1168 | No |
| 117 | Ptcd2 | 16342 | -0.142 | -0.1241 | No |
| 118 | Plvap | 16687 | -0.154 | -0.1350 | No |
| 119 | Glrx | 16805 | -0.159 | -0.1362 | No |
| 120 | Ccser2 | 16843 | -0.160 | -0.1339 | No |
| 121 | Cbr4 | 16892 | -0.162 | -0.1320 | No |
| 122 | Ccnd2 | 16953 | -0.164 | -0.1306 | No |
| 123 | Fbxo4 | 17034 | -0.167 | -0.1300 | No |
| 124 | Scn1b | 17164 | -0.172 | -0.1314 | No |
| 125 | Snap91 | 17225 | -0.174 | -0.1297 | No |
| 126 | Emp1 | 17286 | -0.176 | -0.1280 | No |
| 127 | Lat2 | 17423 | -0.182 | -0.1294 | No |
| 128 | Btbd3 | 17443 | -0.183 | -0.1258 | No |
| 129 | Cxcr4 | 17449 | -0.184 | -0.1216 | No |
| 130 | Prrx1 | 17644 | -0.190 | -0.1253 | No |
| 131 | Pcp4 | 17929 | -0.201 | -0.1325 | No |
| 132 | Gng11 | 18063 | -0.206 | -0.1332 | No |
| 133 | Tor1aip2 | 18096 | -0.208 | -0.1296 | No |
| 134 | Gucy1a1 | 18377 | -0.219 | -0.1362 | No |
| 135 | Klf4 | 18455 | -0.223 | -0.1341 | No |
| 136 | Zfp639 | 18747 | -0.233 | -0.1409 | No |
| 137 | Ephb2 | 18805 | -0.236 | -0.1376 | No |
| 138 | Ero1a | 18831 | -0.237 | -0.1330 | No |
| 139 | Rbm4 | 18859 | -0.238 | -0.1284 | No |
| 140 | Ets1 | 19010 | -0.245 | -0.1288 | No |
| 141 | Gabra3 | 19042 | -0.247 | -0.1242 | No |
| 142 | Aldh1a3 | 19103 | -0.249 | -0.1207 | No |
| 143 | Galnt3 | 19202 | -0.255 | -0.1188 | No |
| 144 | Prelid3b | 19300 | -0.259 | -0.1166 | No |
| 145 | Prdm1 | 19370 | -0.263 | -0.1132 | No |
| 146 | Psmb8 | 19489 | -0.270 | -0.1117 | No |
| 147 | G0s2 | 19805 | -0.284 | -0.1183 | No |
| 148 | Abcb1a | 19884 | -0.289 | -0.1147 | No |
| 149 | Arg1 | 20045 | -0.296 | -0.1143 | No |
| 150 | Epb41l3 | 20159 | -0.303 | -0.1118 | No |
| 151 | Tspan13 | 20214 | -0.306 | -0.1067 | No |
| 152 | Ctss | 20305 | -0.309 | -0.1031 | No |
| 153 | Plek2 | 20447 | -0.318 | -0.1015 | No |
| 154 | Tmem176a | 20861 | -0.343 | -0.1108 | No |
| 155 | Sparcl1 | 21110 | -0.358 | -0.1127 | No |
| 156 | Inhba | 21192 | -0.364 | -0.1074 | No |
| 157 | Spp1 | 21369 | -0.378 | -0.1058 | No |
| 158 | Tmem176b | 21445 | -0.383 | -0.0997 | No |
| 159 | Tmem100 | 21514 | -0.386 | -0.0933 | No |
| 160 | Yrdc | 21666 | -0.397 | -0.0901 | No |
| 161 | Gadd45g | 21669 | -0.397 | -0.0806 | No |
| 162 | Trib2 | 21723 | -0.401 | -0.0732 | No |
| 163 | Spon1 | 21833 | -0.410 | -0.0679 | No |
| 164 | Pdcd1lg2 | 21944 | -0.421 | -0.0625 | No |
| 165 | Ptprr | 21959 | -0.422 | -0.0529 | No |
| 166 | Etv4 | 22064 | -0.431 | -0.0469 | No |
| 167 | Anxa10 | 22348 | -0.462 | -0.0478 | No |
| 168 | St6gal1 | 22432 | -0.471 | -0.0400 | No |
| 169 | Gpnmb | 22492 | -0.481 | -0.0309 | No |
| 170 | Aldh1a2 | 22692 | -0.508 | -0.0271 | No |
| 171 | Car2 | 23015 | -0.561 | -0.0273 | No |
| 172 | Il2rg | 23317 | -0.653 | -0.0244 | No |
| 173 | Angptl4 | 23487 | -0.751 | -0.0135 | No |
| 174 | Cmklr1 | 23526 | -0.798 | 0.0042 | No |