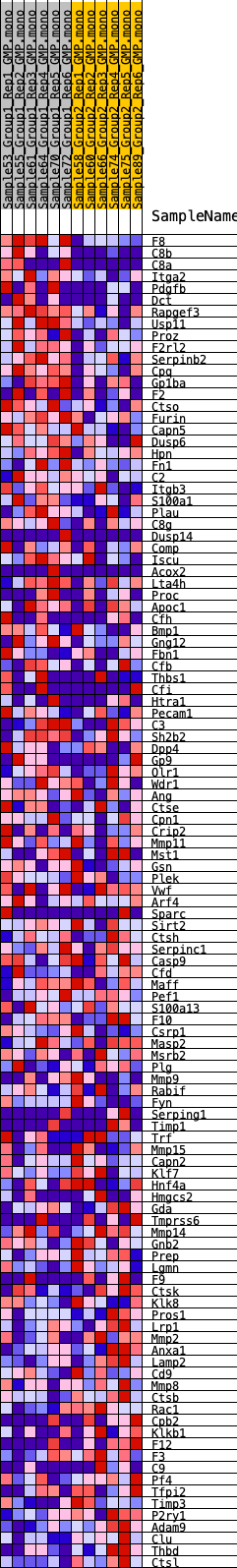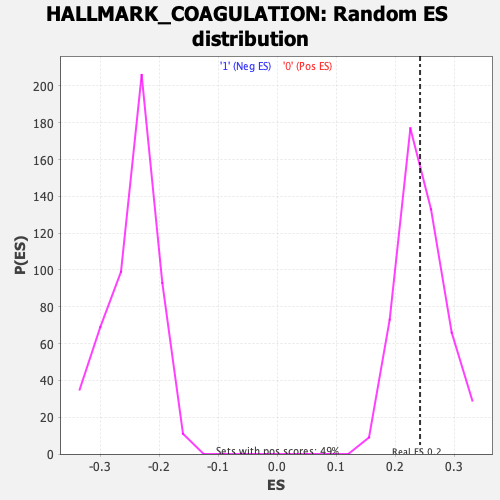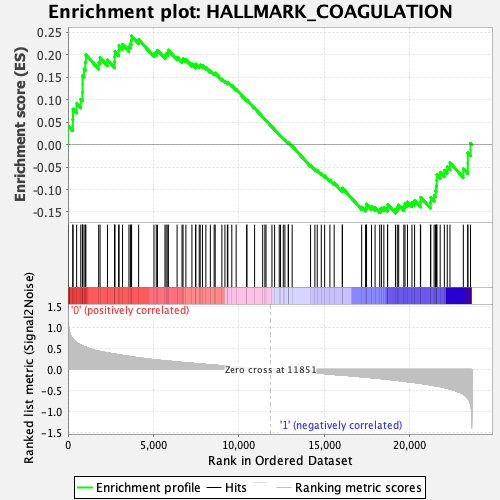
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group2.GMP.mono_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.24216811 |
| Normalized Enrichment Score (NES) | 0.9961071 |
| Nominal p-value | 0.47022587 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.98 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | F8 | 8 | 1.263 | 0.0425 | Yes |
| 2 | C8b | 281 | 0.725 | 0.0554 | Yes |
| 3 | C8a | 293 | 0.722 | 0.0794 | Yes |
| 4 | Itga2 | 505 | 0.633 | 0.0919 | Yes |
| 5 | Pdgfb | 748 | 0.580 | 0.1012 | Yes |
| 6 | Dct | 843 | 0.561 | 0.1162 | Yes |
| 7 | Rapgef3 | 850 | 0.559 | 0.1349 | Yes |
| 8 | Usp11 | 851 | 0.559 | 0.1538 | Yes |
| 9 | Proz | 939 | 0.544 | 0.1686 | Yes |
| 10 | F2rl2 | 1014 | 0.533 | 0.1835 | Yes |
| 11 | Serpinb2 | 1045 | 0.529 | 0.2001 | Yes |
| 12 | Cpq | 1795 | 0.432 | 0.1829 | Yes |
| 13 | Gp1ba | 1875 | 0.426 | 0.1939 | Yes |
| 14 | F2 | 2309 | 0.393 | 0.1888 | Yes |
| 15 | Ctso | 2723 | 0.365 | 0.1836 | Yes |
| 16 | Furin | 2729 | 0.364 | 0.1958 | Yes |
| 17 | Capn5 | 2743 | 0.363 | 0.2075 | Yes |
| 18 | Dusp6 | 2965 | 0.348 | 0.2099 | Yes |
| 19 | Hpn | 2986 | 0.346 | 0.2208 | Yes |
| 20 | Fn1 | 3190 | 0.333 | 0.2234 | Yes |
| 21 | C2 | 3566 | 0.310 | 0.2180 | Yes |
| 22 | Itgb3 | 3650 | 0.304 | 0.2247 | Yes |
| 23 | S100a1 | 3703 | 0.300 | 0.2327 | Yes |
| 24 | Plau | 3720 | 0.300 | 0.2422 | Yes |
| 25 | C8g | 4132 | 0.275 | 0.2340 | No |
| 26 | Dusp14 | 5029 | 0.231 | 0.2037 | No |
| 27 | Comp | 5164 | 0.226 | 0.2057 | No |
| 28 | Iscu | 5232 | 0.222 | 0.2103 | No |
| 29 | Acox2 | 5674 | 0.204 | 0.1985 | No |
| 30 | Lta4h | 5742 | 0.203 | 0.2025 | No |
| 31 | Proc | 5842 | 0.199 | 0.2051 | No |
| 32 | Apoc1 | 5869 | 0.198 | 0.2106 | No |
| 33 | Cfh | 6382 | 0.178 | 0.1949 | No |
| 34 | Bmp1 | 6671 | 0.166 | 0.1883 | No |
| 35 | Gng12 | 6730 | 0.164 | 0.1914 | No |
| 36 | Fbn1 | 6896 | 0.159 | 0.1897 | No |
| 37 | Cfb | 7250 | 0.147 | 0.1797 | No |
| 38 | Thbs1 | 7464 | 0.140 | 0.1754 | No |
| 39 | Cfi | 7479 | 0.140 | 0.1795 | No |
| 40 | Htra1 | 7677 | 0.133 | 0.1756 | No |
| 41 | Pecam1 | 7734 | 0.131 | 0.1777 | No |
| 42 | C3 | 7866 | 0.126 | 0.1764 | No |
| 43 | Sh2b2 | 8065 | 0.119 | 0.1720 | No |
| 44 | Dpp4 | 8327 | 0.110 | 0.1646 | No |
| 45 | Gp9 | 8551 | 0.102 | 0.1586 | No |
| 46 | Olr1 | 8611 | 0.100 | 0.1595 | No |
| 47 | Wdr1 | 9003 | 0.089 | 0.1459 | No |
| 48 | Ang | 9181 | 0.083 | 0.1411 | No |
| 49 | Ctse | 9325 | 0.079 | 0.1377 | No |
| 50 | Cpn1 | 9352 | 0.078 | 0.1392 | No |
| 51 | Crip2 | 9583 | 0.070 | 0.1318 | No |
| 52 | Mmp11 | 9837 | 0.062 | 0.1232 | No |
| 53 | Mst1 | 10453 | 0.043 | 0.0985 | No |
| 54 | Gsn | 10466 | 0.043 | 0.0994 | No |
| 55 | Plek | 10917 | 0.029 | 0.0813 | No |
| 56 | Vwf | 11389 | 0.013 | 0.0617 | No |
| 57 | Arf4 | 11518 | 0.009 | 0.0565 | No |
| 58 | Sparc | 11584 | 0.008 | 0.0540 | No |
| 59 | Sirt2 | 11932 | -0.002 | 0.0393 | No |
| 60 | Ctsh | 12088 | -0.007 | 0.0330 | No |
| 61 | Serpinc1 | 12358 | -0.015 | 0.0220 | No |
| 62 | Casp9 | 12430 | -0.017 | 0.0196 | No |
| 63 | Cfd | 12594 | -0.022 | 0.0134 | No |
| 64 | Maff | 12688 | -0.025 | 0.0103 | No |
| 65 | Pef1 | 12890 | -0.031 | 0.0028 | No |
| 66 | S100a13 | 12892 | -0.032 | 0.0038 | No |
| 67 | F10 | 12897 | -0.032 | 0.0047 | No |
| 68 | Csrp1 | 13114 | -0.039 | -0.0031 | No |
| 69 | Masp2 | 14191 | -0.072 | -0.0464 | No |
| 70 | Msrb2 | 14442 | -0.080 | -0.0544 | No |
| 71 | Plg | 14580 | -0.084 | -0.0574 | No |
| 72 | Mmp9 | 14818 | -0.091 | -0.0644 | No |
| 73 | Rabif | 15015 | -0.098 | -0.0694 | No |
| 74 | Fyn | 15322 | -0.109 | -0.0787 | No |
| 75 | Serping1 | 15571 | -0.118 | -0.0853 | No |
| 76 | Timp1 | 16047 | -0.132 | -0.1010 | No |
| 77 | Trf | 16053 | -0.132 | -0.0967 | No |
| 78 | Mmp15 | 17180 | -0.172 | -0.1388 | No |
| 79 | Capn2 | 17409 | -0.182 | -0.1423 | No |
| 80 | Klf7 | 17455 | -0.184 | -0.1380 | No |
| 81 | Hnf4a | 17465 | -0.184 | -0.1322 | No |
| 82 | Hmgcs2 | 17756 | -0.194 | -0.1379 | No |
| 83 | Gda | 17965 | -0.202 | -0.1399 | No |
| 84 | Tmprss6 | 18236 | -0.213 | -0.1442 | No |
| 85 | Mmp14 | 18336 | -0.217 | -0.1410 | No |
| 86 | Gnb2 | 18478 | -0.224 | -0.1394 | No |
| 87 | Prep | 18691 | -0.231 | -0.1406 | No |
| 88 | Lgmn | 18702 | -0.231 | -0.1332 | No |
| 89 | F9 | 19158 | -0.253 | -0.1440 | No |
| 90 | Ctsk | 19264 | -0.258 | -0.1397 | No |
| 91 | Klk8 | 19346 | -0.262 | -0.1343 | No |
| 92 | Pros1 | 19647 | -0.277 | -0.1377 | No |
| 93 | Lrp1 | 19716 | -0.281 | -0.1311 | No |
| 94 | Mmp2 | 19861 | -0.287 | -0.1274 | No |
| 95 | Anxa1 | 20123 | -0.301 | -0.1283 | No |
| 96 | Lamp2 | 20277 | -0.308 | -0.1244 | No |
| 97 | Cd9 | 20616 | -0.328 | -0.1277 | No |
| 98 | Mmp8 | 20630 | -0.329 | -0.1171 | No |
| 99 | Ctsb | 21210 | -0.366 | -0.1293 | No |
| 100 | Rac1 | 21223 | -0.366 | -0.1175 | No |
| 101 | Cpb2 | 21420 | -0.382 | -0.1129 | No |
| 102 | Klkb1 | 21503 | -0.386 | -0.1033 | No |
| 103 | F12 | 21540 | -0.388 | -0.0917 | No |
| 104 | F3 | 21572 | -0.391 | -0.0797 | No |
| 105 | C9 | 21577 | -0.391 | -0.0667 | No |
| 106 | Pf4 | 21773 | -0.406 | -0.0612 | No |
| 107 | Tfpi2 | 22017 | -0.428 | -0.0570 | No |
| 108 | Timp3 | 22187 | -0.444 | -0.0492 | No |
| 109 | P2ry1 | 22346 | -0.462 | -0.0403 | No |
| 110 | Adam9 | 23128 | -0.589 | -0.0535 | No |
| 111 | Clu | 23389 | -0.683 | -0.0414 | No |
| 112 | Thbd | 23391 | -0.684 | -0.0183 | No |
| 113 | Ctsl | 23548 | -0.831 | 0.0032 | No |