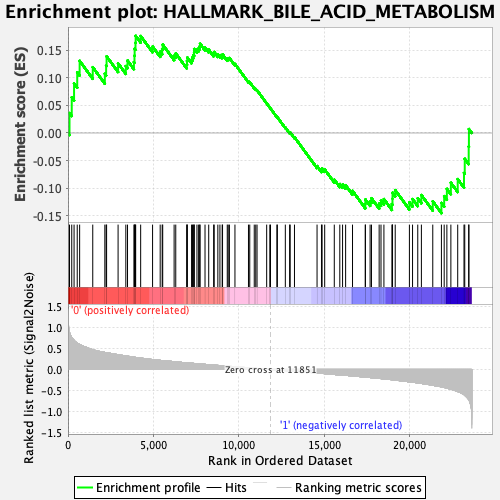
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group2.GMP.mono_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.17620511 |
| Normalized Enrichment Score (NES) | 0.7793996 |
| Nominal p-value | 0.8795181 |
| FDR q-value | 0.83458143 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abca9 | 88 | 0.888 | 0.0362 | Yes |
| 2 | Hsd3b7 | 219 | 0.755 | 0.0646 | Yes |
| 3 | Abca6 | 355 | 0.691 | 0.0900 | Yes |
| 4 | Gnmt | 540 | 0.623 | 0.1101 | Yes |
| 5 | Dio1 | 681 | 0.593 | 0.1309 | Yes |
| 6 | Nudt12 | 1448 | 0.469 | 0.1194 | Yes |
| 7 | Cyp39a1 | 2155 | 0.404 | 0.1075 | Yes |
| 8 | Hao1 | 2231 | 0.399 | 0.1223 | Yes |
| 9 | Nedd4 | 2258 | 0.397 | 0.1391 | Yes |
| 10 | Abca3 | 2930 | 0.351 | 0.1263 | Yes |
| 11 | Abcd2 | 3383 | 0.321 | 0.1215 | Yes |
| 12 | Pex26 | 3482 | 0.315 | 0.1315 | Yes |
| 13 | Atxn1 | 3857 | 0.291 | 0.1287 | Yes |
| 14 | Abca2 | 3889 | 0.289 | 0.1404 | Yes |
| 15 | Acsl1 | 3899 | 0.289 | 0.1530 | Yes |
| 16 | Nr1h4 | 3948 | 0.286 | 0.1638 | Yes |
| 17 | Abcd1 | 3959 | 0.286 | 0.1762 | Yes |
| 18 | Pnpla8 | 4251 | 0.269 | 0.1759 | No |
| 19 | Hacl1 | 4948 | 0.234 | 0.1569 | No |
| 20 | Pex11g | 5393 | 0.216 | 0.1477 | No |
| 21 | Nr3c2 | 5510 | 0.210 | 0.1522 | No |
| 22 | Bmp6 | 5541 | 0.209 | 0.1604 | No |
| 23 | Pex7 | 6208 | 0.184 | 0.1403 | No |
| 24 | Rbp1 | 6304 | 0.181 | 0.1444 | No |
| 25 | Pex1 | 6948 | 0.157 | 0.1242 | No |
| 26 | Pfkm | 6963 | 0.156 | 0.1306 | No |
| 27 | Ar | 6980 | 0.156 | 0.1369 | No |
| 28 | Cyp27a1 | 7229 | 0.147 | 0.1330 | No |
| 29 | Hsd17b11 | 7282 | 0.146 | 0.1374 | No |
| 30 | Slc27a2 | 7334 | 0.144 | 0.1417 | No |
| 31 | Fdxr | 7379 | 0.143 | 0.1462 | No |
| 32 | Idh1 | 7384 | 0.143 | 0.1525 | No |
| 33 | Lonp2 | 7544 | 0.137 | 0.1519 | No |
| 34 | Efhc1 | 7642 | 0.134 | 0.1538 | No |
| 35 | Pex13 | 7703 | 0.132 | 0.1572 | No |
| 36 | Slc23a1 | 7727 | 0.131 | 0.1621 | No |
| 37 | Mlycd | 8016 | 0.121 | 0.1553 | No |
| 38 | Phyh | 8232 | 0.113 | 0.1512 | No |
| 39 | Crot | 8521 | 0.103 | 0.1436 | No |
| 40 | Npc1 | 8552 | 0.102 | 0.1469 | No |
| 41 | Gnpat | 8766 | 0.096 | 0.1422 | No |
| 42 | Aldh1a1 | 8888 | 0.092 | 0.1412 | No |
| 43 | Gstk1 | 9020 | 0.088 | 0.1396 | No |
| 44 | Cyp46a1 | 9041 | 0.087 | 0.1427 | No |
| 45 | Sult2b1 | 9317 | 0.079 | 0.1345 | No |
| 46 | Slc27a5 | 9373 | 0.077 | 0.1356 | No |
| 47 | Pex11a | 9443 | 0.074 | 0.1361 | No |
| 48 | Cyp7b1 | 9767 | 0.065 | 0.1252 | No |
| 49 | Abca5 | 10558 | 0.039 | 0.0934 | No |
| 50 | Pecr | 10633 | 0.037 | 0.0919 | No |
| 51 | Hsd17b4 | 10896 | 0.029 | 0.0821 | No |
| 52 | Slc22a18 | 10984 | 0.026 | 0.0796 | No |
| 53 | Paox | 11059 | 0.024 | 0.0775 | No |
| 54 | Slc23a2 | 11623 | 0.006 | 0.0539 | No |
| 55 | Pex6 | 11807 | 0.001 | 0.0462 | No |
| 56 | Nr0b2 | 11847 | 0.000 | 0.0445 | No |
| 57 | Pex19 | 12220 | -0.011 | 0.0292 | No |
| 58 | Pex12 | 12249 | -0.012 | 0.0285 | No |
| 59 | Abca4 | 12714 | -0.026 | 0.0100 | No |
| 60 | Lipe | 12966 | -0.034 | 0.0009 | No |
| 61 | Amacr | 12997 | -0.035 | 0.0012 | No |
| 62 | Pipox | 13249 | -0.043 | -0.0076 | No |
| 63 | Cat | 14570 | -0.083 | -0.0599 | No |
| 64 | Abca8b | 14829 | -0.091 | -0.0668 | No |
| 65 | Gc | 14865 | -0.093 | -0.0641 | No |
| 66 | Abcd3 | 15017 | -0.098 | -0.0661 | No |
| 67 | Ephx2 | 15578 | -0.118 | -0.0846 | No |
| 68 | Dio2 | 15901 | -0.127 | -0.0926 | No |
| 69 | Aldh9a1 | 16063 | -0.133 | -0.0934 | No |
| 70 | Slc29a1 | 16242 | -0.139 | -0.0948 | No |
| 71 | Optn | 16645 | -0.153 | -0.1050 | No |
| 72 | Akr1d1 | 17393 | -0.181 | -0.1286 | No |
| 73 | Bcar3 | 17397 | -0.181 | -0.1206 | No |
| 74 | Pxmp2 | 17674 | -0.191 | -0.1237 | No |
| 75 | Abca1 | 17754 | -0.194 | -0.1183 | No |
| 76 | Tfcp2l1 | 18203 | -0.212 | -0.1279 | No |
| 77 | Prdx5 | 18311 | -0.216 | -0.1227 | No |
| 78 | Acsl5 | 18482 | -0.224 | -0.1199 | No |
| 79 | Slc35b2 | 18950 | -0.242 | -0.1288 | No |
| 80 | Dhcr24 | 18983 | -0.244 | -0.1192 | No |
| 81 | Idh2 | 18986 | -0.244 | -0.1083 | No |
| 82 | Idi1 | 19143 | -0.252 | -0.1036 | No |
| 83 | Rxra | 19973 | -0.292 | -0.1257 | No |
| 84 | Pex16 | 20157 | -0.303 | -0.1199 | No |
| 85 | Isoc1 | 20462 | -0.318 | -0.1185 | No |
| 86 | Scp2 | 20674 | -0.331 | -0.1125 | No |
| 87 | Retsat | 21340 | -0.376 | -0.1239 | No |
| 88 | Aqp9 | 21846 | -0.412 | -0.1269 | No |
| 89 | Fads2 | 22008 | -0.427 | -0.1145 | No |
| 90 | Sod1 | 22165 | -0.441 | -0.1013 | No |
| 91 | Klf1 | 22399 | -0.467 | -0.0902 | No |
| 92 | Lck | 22799 | -0.524 | -0.0836 | No |
| 93 | Soat2 | 23167 | -0.596 | -0.0724 | No |
| 94 | Abcg4 | 23211 | -0.610 | -0.0468 | No |
| 95 | Gclm | 23442 | -0.711 | -0.0246 | No |
| 96 | Fads1 | 23455 | -0.718 | 0.0072 | No |