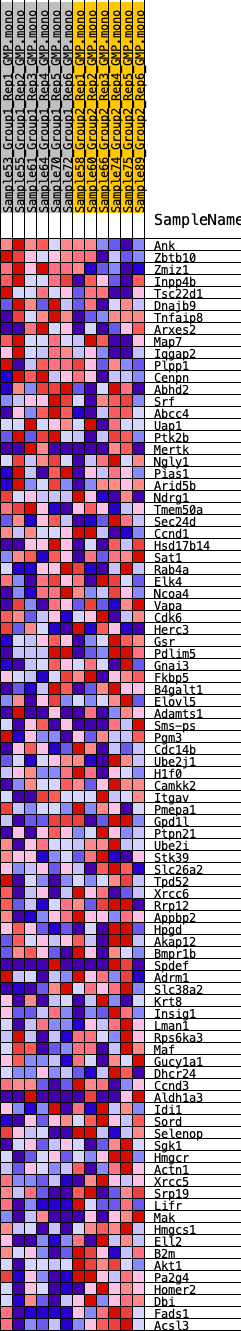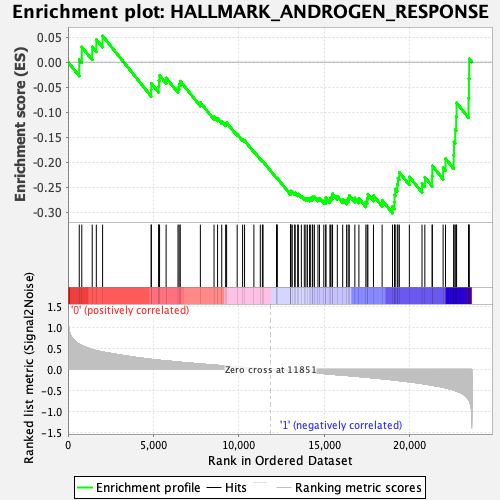
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | GMP.GMP.mono_Pheno.cls #Group1_versus_Group2.GMP.mono_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | GMP.mono_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_ANDROGEN_RESPONSE |
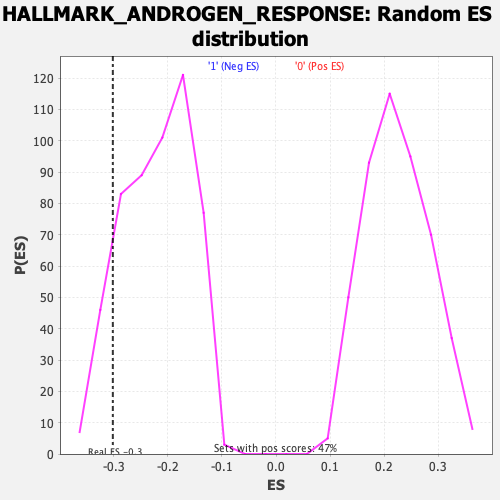
| Enrichment Score (ES) | -0.30152905 |
| Normalized Enrichment Score (NES) | -1.3655907 |
| Nominal p-value | 0.10246679 |
| FDR q-value | 0.15931259 |
| FWER p-Value | 0.619 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ank | 658 | 0.597 | 0.0050 | No |
| 2 | Zbtb10 | 806 | 0.568 | 0.0302 | No |
| 3 | Zmiz1 | 1417 | 0.474 | 0.0305 | No |
| 4 | Inpp4b | 1657 | 0.446 | 0.0450 | No |
| 5 | Tsc22d1 | 2024 | 0.414 | 0.0523 | No |
| 6 | Dnajb9 | 4869 | 0.238 | -0.0554 | No |
| 7 | Tnfaip8 | 4871 | 0.238 | -0.0422 | No |
| 8 | Arxes2 | 5305 | 0.219 | -0.0486 | No |
| 9 | Map7 | 5322 | 0.218 | -0.0372 | No |
| 10 | Iqgap2 | 5357 | 0.217 | -0.0266 | No |
| 11 | Plpp1 | 5740 | 0.203 | -0.0317 | No |
| 12 | Cenpn | 6435 | 0.176 | -0.0514 | No |
| 13 | Abhd2 | 6502 | 0.174 | -0.0447 | No |
| 14 | Srf | 6567 | 0.171 | -0.0379 | No |
| 15 | Abcc4 | 7744 | 0.130 | -0.0807 | No |
| 16 | Uap1 | 8544 | 0.103 | -0.1090 | No |
| 17 | Ptk2b | 8750 | 0.096 | -0.1124 | No |
| 18 | Mertk | 8995 | 0.089 | -0.1178 | No |
| 19 | Ngly1 | 9223 | 0.082 | -0.1230 | No |
| 20 | Pias1 | 9279 | 0.080 | -0.1209 | No |
| 21 | Arid5b | 9897 | 0.060 | -0.1438 | No |
| 22 | Ndrg1 | 10214 | 0.050 | -0.1544 | No |
| 23 | Tmem50a | 10329 | 0.047 | -0.1566 | No |
| 24 | Sec24d | 10876 | 0.030 | -0.1782 | No |
| 25 | Ccnd1 | 11250 | 0.017 | -0.1931 | No |
| 26 | Hsd17b14 | 11373 | 0.014 | -0.1975 | No |
| 27 | Sat1 | 11411 | 0.012 | -0.1984 | No |
| 28 | Rab4a | 12201 | -0.010 | -0.2313 | No |
| 29 | Elk4 | 12212 | -0.011 | -0.2312 | No |
| 30 | Ncoa4 | 12245 | -0.012 | -0.2319 | No |
| 31 | Vapa | 13015 | -0.036 | -0.2626 | No |
| 32 | Cdk6 | 13024 | -0.036 | -0.2609 | No |
| 33 | Herc3 | 13033 | -0.036 | -0.2593 | No |
| 34 | Gsr | 13035 | -0.036 | -0.2573 | No |
| 35 | Pdlim5 | 13118 | -0.039 | -0.2586 | No |
| 36 | Gnai3 | 13262 | -0.043 | -0.2623 | No |
| 37 | Fkbp5 | 13288 | -0.044 | -0.2610 | No |
| 38 | B4galt1 | 13438 | -0.049 | -0.2646 | No |
| 39 | Elovl5 | 13476 | -0.050 | -0.2634 | No |
| 40 | Adamts1 | 13651 | -0.056 | -0.2677 | No |
| 41 | Sms-ps | 13825 | -0.061 | -0.2717 | No |
| 42 | Pgm3 | 13915 | -0.064 | -0.2719 | No |
| 43 | Cdc14b | 14005 | -0.067 | -0.2720 | No |
| 44 | Ube2j1 | 14133 | -0.071 | -0.2735 | No |
| 45 | H1f0 | 14177 | -0.072 | -0.2714 | No |
| 46 | Camkk2 | 14295 | -0.075 | -0.2722 | No |
| 47 | Itgav | 14321 | -0.076 | -0.2691 | No |
| 48 | Pmepa1 | 14414 | -0.079 | -0.2687 | No |
| 49 | Gpd1l | 14622 | -0.085 | -0.2728 | No |
| 50 | Ptpn21 | 14711 | -0.088 | -0.2716 | No |
| 51 | Ube2i | 14965 | -0.096 | -0.2771 | No |
| 52 | Stk39 | 15083 | -0.101 | -0.2765 | No |
| 53 | Slc26a2 | 15084 | -0.101 | -0.2709 | No |
| 54 | Tpd52 | 15324 | -0.109 | -0.2751 | No |
| 55 | Xrcc6 | 15365 | -0.110 | -0.2707 | No |
| 56 | Rrp12 | 15448 | -0.113 | -0.2679 | No |
| 57 | Appbp2 | 15478 | -0.114 | -0.2628 | No |
| 58 | Hpgd | 15756 | -0.124 | -0.2678 | No |
| 59 | Akap12 | 16070 | -0.133 | -0.2737 | No |
| 60 | Bmpr1b | 16300 | -0.141 | -0.2757 | No |
| 61 | Spdef | 16390 | -0.144 | -0.2715 | No |
| 62 | Adrm1 | 16456 | -0.146 | -0.2662 | No |
| 63 | Slc38a2 | 16782 | -0.158 | -0.2713 | No |
| 64 | Krt8 | 17019 | -0.167 | -0.2721 | No |
| 65 | Insig1 | 17425 | -0.182 | -0.2792 | No |
| 66 | Lman1 | 17503 | -0.185 | -0.2722 | No |
| 67 | Rps6ka3 | 17544 | -0.187 | -0.2636 | No |
| 68 | Maf | 17871 | -0.199 | -0.2665 | No |
| 69 | Gucy1a1 | 18377 | -0.219 | -0.2758 | No |
| 70 | Dhcr24 | 18983 | -0.244 | -0.2881 | Yes |
| 71 | Ccnd3 | 19099 | -0.249 | -0.2792 | Yes |
| 72 | Aldh1a3 | 19103 | -0.249 | -0.2655 | Yes |
| 73 | Idi1 | 19143 | -0.252 | -0.2533 | Yes |
| 74 | Sord | 19263 | -0.258 | -0.2441 | Yes |
| 75 | Selenop | 19304 | -0.260 | -0.2314 | Yes |
| 76 | Sgk1 | 19385 | -0.264 | -0.2202 | Yes |
| 77 | Hmgcr | 19970 | -0.292 | -0.2289 | Yes |
| 78 | Actn1 | 20711 | -0.334 | -0.2419 | Yes |
| 79 | Xrcc5 | 20876 | -0.344 | -0.2299 | Yes |
| 80 | Srp19 | 21303 | -0.373 | -0.2274 | Yes |
| 81 | Lifr | 21314 | -0.373 | -0.2072 | Yes |
| 82 | Mak | 21945 | -0.421 | -0.2107 | Yes |
| 83 | Hmgcs1 | 22086 | -0.434 | -0.1927 | Yes |
| 84 | Ell2 | 22564 | -0.491 | -0.1859 | Yes |
| 85 | B2m | 22580 | -0.492 | -0.1593 | Yes |
| 86 | Akt1 | 22654 | -0.502 | -0.1347 | Yes |
| 87 | Pa2g4 | 22710 | -0.510 | -0.1088 | Yes |
| 88 | Homer2 | 22734 | -0.515 | -0.0814 | Yes |
| 89 | Dbi | 23434 | -0.707 | -0.0720 | Yes |
| 90 | Fads1 | 23455 | -0.718 | -0.0332 | Yes |
| 91 | Acsl3 | 23471 | -0.730 | 0.0065 | Yes |