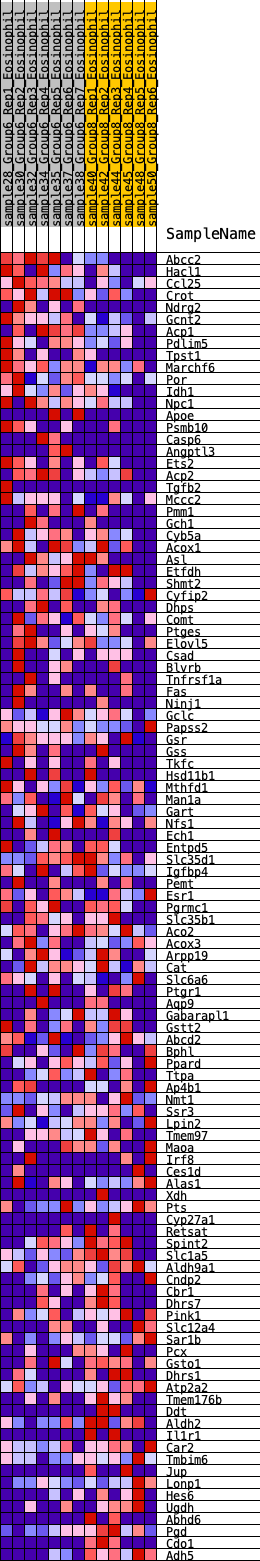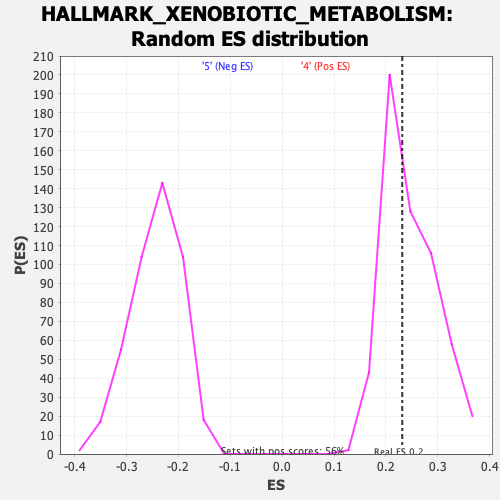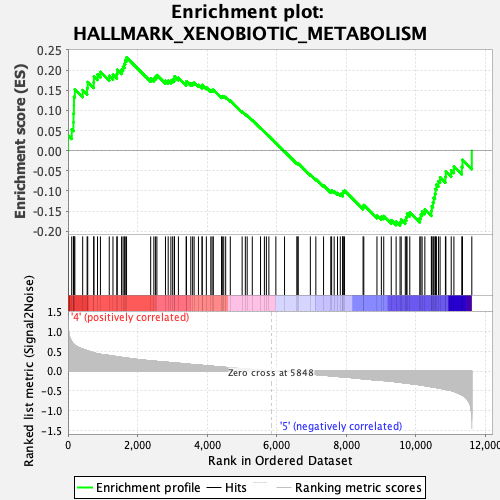
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group6_versus_Group8.Eosinophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Eosinophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | 0.23151743 |
| Normalized Enrichment Score (NES) | 0.9353606 |
| Nominal p-value | 0.53859967 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abcc2 | 3 | 1.213 | 0.0381 | Yes |
| 2 | Hacl1 | 107 | 0.749 | 0.0528 | Yes |
| 3 | Ccl25 | 156 | 0.690 | 0.0704 | Yes |
| 4 | Crot | 159 | 0.687 | 0.0920 | Yes |
| 5 | Ndrg2 | 170 | 0.677 | 0.1125 | Yes |
| 6 | Gcnt2 | 172 | 0.675 | 0.1337 | Yes |
| 7 | Acp1 | 197 | 0.654 | 0.1523 | Yes |
| 8 | Pdlim5 | 422 | 0.557 | 0.1504 | Yes |
| 9 | Tpst1 | 550 | 0.518 | 0.1558 | Yes |
| 10 | Marchf6 | 567 | 0.511 | 0.1705 | Yes |
| 11 | Por | 740 | 0.463 | 0.1702 | Yes |
| 12 | Idh1 | 745 | 0.462 | 0.1844 | Yes |
| 13 | Npc1 | 848 | 0.437 | 0.1893 | Yes |
| 14 | Apoe | 932 | 0.424 | 0.1955 | Yes |
| 15 | Psmb10 | 1186 | 0.392 | 0.1859 | Yes |
| 16 | Casp6 | 1293 | 0.378 | 0.1886 | Yes |
| 17 | Angptl3 | 1404 | 0.362 | 0.1905 | Yes |
| 18 | Ets2 | 1415 | 0.361 | 0.2011 | Yes |
| 19 | Acp2 | 1540 | 0.346 | 0.2012 | Yes |
| 20 | Tgfb2 | 1583 | 0.341 | 0.2084 | Yes |
| 21 | Mccc2 | 1621 | 0.337 | 0.2158 | Yes |
| 22 | Pmm1 | 1645 | 0.334 | 0.2243 | Yes |
| 23 | Gch1 | 1683 | 0.329 | 0.2315 | Yes |
| 24 | Cyb5a | 2377 | 0.259 | 0.1794 | No |
| 25 | Acox1 | 2468 | 0.251 | 0.1795 | No |
| 26 | Asl | 2513 | 0.248 | 0.1836 | No |
| 27 | Etfdh | 2555 | 0.244 | 0.1877 | No |
| 28 | Shmt2 | 2802 | 0.226 | 0.1735 | No |
| 29 | Cyfip2 | 2880 | 0.220 | 0.1737 | No |
| 30 | Dhps | 2958 | 0.213 | 0.1738 | No |
| 31 | Comt | 3009 | 0.209 | 0.1760 | No |
| 32 | Ptges | 3056 | 0.205 | 0.1785 | No |
| 33 | Elovl5 | 3062 | 0.204 | 0.1845 | No |
| 34 | Csad | 3174 | 0.195 | 0.1810 | No |
| 35 | Blvrb | 3397 | 0.179 | 0.1674 | No |
| 36 | Tnfrsf1a | 3405 | 0.179 | 0.1724 | No |
| 37 | Fas | 3531 | 0.168 | 0.1669 | No |
| 38 | Ninj1 | 3583 | 0.164 | 0.1676 | No |
| 39 | Gclc | 3626 | 0.162 | 0.1691 | No |
| 40 | Papss2 | 3750 | 0.152 | 0.1632 | No |
| 41 | Gsr | 3855 | 0.145 | 0.1587 | No |
| 42 | Gss | 3857 | 0.145 | 0.1632 | No |
| 43 | Tkfc | 3977 | 0.135 | 0.1571 | No |
| 44 | Hsd11b1 | 4106 | 0.124 | 0.1499 | No |
| 45 | Mthfd1 | 4142 | 0.122 | 0.1507 | No |
| 46 | Man1a | 4178 | 0.119 | 0.1514 | No |
| 47 | Gart | 4414 | 0.101 | 0.1342 | No |
| 48 | Nfs1 | 4436 | 0.099 | 0.1355 | No |
| 49 | Ech1 | 4477 | 0.097 | 0.1351 | No |
| 50 | Entpd5 | 4533 | 0.092 | 0.1332 | No |
| 51 | Slc35d1 | 4666 | 0.083 | 0.1244 | No |
| 52 | Igfbp4 | 5006 | 0.058 | 0.0967 | No |
| 53 | Pemt | 5098 | 0.052 | 0.0904 | No |
| 54 | Esr1 | 5151 | 0.049 | 0.0875 | No |
| 55 | Pgrmc1 | 5298 | 0.038 | 0.0760 | No |
| 56 | Slc35b1 | 5535 | 0.020 | 0.0561 | No |
| 57 | Aco2 | 5646 | 0.013 | 0.0469 | No |
| 58 | Acox3 | 5703 | 0.008 | 0.0423 | No |
| 59 | Arpp19 | 5775 | 0.004 | 0.0363 | No |
| 60 | Cat | 5976 | -0.000 | 0.0189 | No |
| 61 | Slc6a6 | 6225 | -0.019 | -0.0021 | No |
| 62 | Ptgr1 | 6578 | -0.045 | -0.0313 | No |
| 63 | Aqp9 | 6608 | -0.047 | -0.0323 | No |
| 64 | Gabarapl1 | 6616 | -0.048 | -0.0314 | No |
| 65 | Gstt2 | 6967 | -0.074 | -0.0595 | No |
| 66 | Abcd2 | 7125 | -0.086 | -0.0704 | No |
| 67 | Bphl | 7347 | -0.103 | -0.0864 | No |
| 68 | Ppard | 7555 | -0.120 | -0.1006 | No |
| 69 | Ttpa | 7579 | -0.121 | -0.0988 | No |
| 70 | Ap4b1 | 7652 | -0.126 | -0.1011 | No |
| 71 | Nmt1 | 7748 | -0.134 | -0.1051 | No |
| 72 | Ssr3 | 7828 | -0.141 | -0.1075 | No |
| 73 | Lpin2 | 7897 | -0.146 | -0.1088 | No |
| 74 | Tmem97 | 7901 | -0.146 | -0.1045 | No |
| 75 | Maoa | 7912 | -0.147 | -0.1007 | No |
| 76 | Irf8 | 7950 | -0.149 | -0.0992 | No |
| 77 | Ces1d | 8483 | -0.194 | -0.1393 | No |
| 78 | Alas1 | 8504 | -0.196 | -0.1349 | No |
| 79 | Xdh | 8882 | -0.228 | -0.1604 | No |
| 80 | Pts | 9009 | -0.237 | -0.1639 | No |
| 81 | Cyp27a1 | 9077 | -0.239 | -0.1622 | No |
| 82 | Retsat | 9293 | -0.256 | -0.1728 | No |
| 83 | Spint2 | 9433 | -0.272 | -0.1763 | No |
| 84 | Slc1a5 | 9546 | -0.282 | -0.1771 | No |
| 85 | Aldh9a1 | 9579 | -0.287 | -0.1708 | No |
| 86 | Cndp2 | 9697 | -0.300 | -0.1715 | No |
| 87 | Cbr1 | 9727 | -0.302 | -0.1645 | No |
| 88 | Dhrs7 | 9747 | -0.306 | -0.1565 | No |
| 89 | Pink1 | 9826 | -0.313 | -0.1534 | No |
| 90 | Slc12a4 | 10113 | -0.345 | -0.1673 | No |
| 91 | Sar1b | 10144 | -0.350 | -0.1589 | No |
| 92 | Pcx | 10180 | -0.356 | -0.1507 | No |
| 93 | Gsto1 | 10259 | -0.367 | -0.1459 | No |
| 94 | Dhrs1 | 10446 | -0.395 | -0.1495 | No |
| 95 | Atp2a2 | 10457 | -0.398 | -0.1378 | No |
| 96 | Tmem176b | 10492 | -0.404 | -0.1280 | No |
| 97 | Ddt | 10514 | -0.406 | -0.1170 | No |
| 98 | Aldh2 | 10550 | -0.410 | -0.1071 | No |
| 99 | Il1r1 | 10565 | -0.413 | -0.0953 | No |
| 100 | Car2 | 10591 | -0.416 | -0.0843 | No |
| 101 | Tmbim6 | 10650 | -0.427 | -0.0759 | No |
| 102 | Jup | 10697 | -0.436 | -0.0661 | No |
| 103 | Lonp1 | 10850 | -0.461 | -0.0648 | No |
| 104 | Hes6 | 10863 | -0.463 | -0.0512 | No |
| 105 | Ugdh | 11017 | -0.492 | -0.0489 | No |
| 106 | Abhd6 | 11094 | -0.515 | -0.0393 | No |
| 107 | Pgd | 11321 | -0.595 | -0.0401 | No |
| 108 | Cdo1 | 11341 | -0.606 | -0.0226 | No |
| 109 | Adh5 | 11609 | -1.451 | 0.0000 | No |