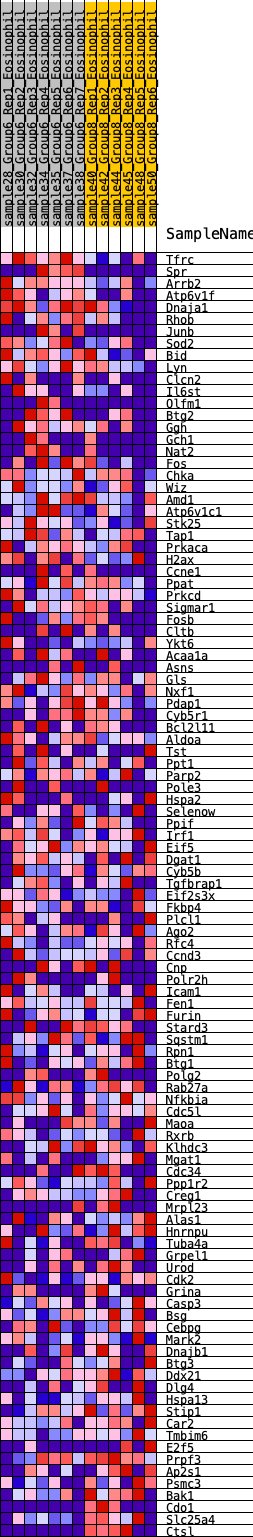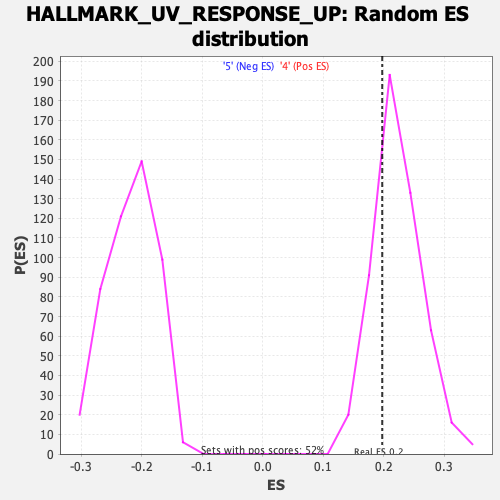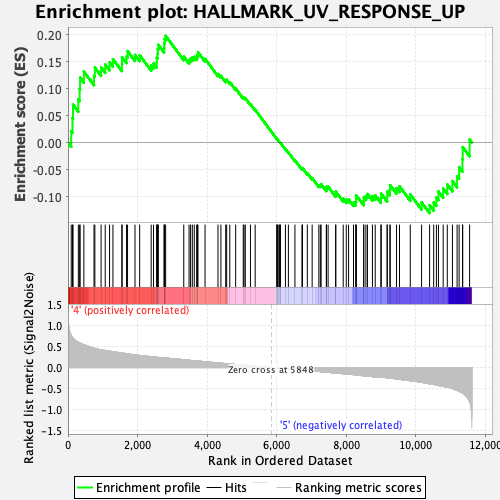
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group6_versus_Group8.Eosinophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Eosinophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | 0.19768229 |
| Normalized Enrichment Score (NES) | 0.8883166 |
| Nominal p-value | 0.76199615 |
| FDR q-value | 0.9570891 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tfrc | 93 | 0.770 | 0.0211 | Yes |
| 2 | Spr | 129 | 0.715 | 0.0452 | Yes |
| 3 | Arrb2 | 145 | 0.696 | 0.0703 | Yes |
| 4 | Atp6v1f | 297 | 0.602 | 0.0800 | Yes |
| 5 | Dnaja1 | 335 | 0.588 | 0.0991 | Yes |
| 6 | Rhob | 348 | 0.583 | 0.1202 | Yes |
| 7 | Junb | 456 | 0.545 | 0.1315 | Yes |
| 8 | Sod2 | 748 | 0.460 | 0.1237 | Yes |
| 9 | Bid | 772 | 0.455 | 0.1390 | Yes |
| 10 | Lyn | 951 | 0.421 | 0.1395 | Yes |
| 11 | Clcn2 | 1070 | 0.407 | 0.1447 | Yes |
| 12 | Il6st | 1195 | 0.390 | 0.1487 | Yes |
| 13 | Olfm1 | 1292 | 0.378 | 0.1547 | Yes |
| 14 | Btg2 | 1550 | 0.345 | 0.1454 | Yes |
| 15 | Ggh | 1554 | 0.345 | 0.1583 | Yes |
| 16 | Gch1 | 1683 | 0.329 | 0.1596 | Yes |
| 17 | Nat2 | 1712 | 0.326 | 0.1695 | Yes |
| 18 | Fos | 1926 | 0.301 | 0.1624 | Yes |
| 19 | Chka | 2060 | 0.287 | 0.1618 | Yes |
| 20 | Wiz | 2391 | 0.258 | 0.1429 | Yes |
| 21 | Amd1 | 2459 | 0.252 | 0.1466 | Yes |
| 22 | Atp6v1c1 | 2550 | 0.245 | 0.1481 | Yes |
| 23 | Stk25 | 2552 | 0.245 | 0.1573 | Yes |
| 24 | Tap1 | 2575 | 0.242 | 0.1645 | Yes |
| 25 | Prkaca | 2583 | 0.242 | 0.1731 | Yes |
| 26 | H2ax | 2596 | 0.240 | 0.1811 | Yes |
| 27 | Ccne1 | 2763 | 0.229 | 0.1754 | Yes |
| 28 | Ppat | 2767 | 0.229 | 0.1838 | Yes |
| 29 | Prkcd | 2777 | 0.228 | 0.1917 | Yes |
| 30 | Sigmar1 | 2807 | 0.225 | 0.1977 | Yes |
| 31 | Fosb | 3328 | 0.186 | 0.1595 | No |
| 32 | Cltb | 3480 | 0.172 | 0.1529 | No |
| 33 | Ykt6 | 3524 | 0.168 | 0.1556 | No |
| 34 | Acaa1a | 3574 | 0.165 | 0.1576 | No |
| 35 | Asns | 3630 | 0.161 | 0.1589 | No |
| 36 | Gls | 3699 | 0.156 | 0.1589 | No |
| 37 | Nxf1 | 3723 | 0.154 | 0.1628 | No |
| 38 | Pdap1 | 3736 | 0.153 | 0.1675 | No |
| 39 | Cyb5r1 | 3940 | 0.138 | 0.1551 | No |
| 40 | Bcl2l11 | 4312 | 0.109 | 0.1270 | No |
| 41 | Aldoa | 4396 | 0.102 | 0.1236 | No |
| 42 | Tst | 4534 | 0.092 | 0.1152 | No |
| 43 | Ppt1 | 4561 | 0.091 | 0.1164 | No |
| 44 | Parp2 | 4651 | 0.084 | 0.1119 | No |
| 45 | Pole3 | 4820 | 0.071 | 0.1000 | No |
| 46 | Hspa2 | 5038 | 0.056 | 0.0832 | No |
| 47 | Selenow | 5061 | 0.054 | 0.0833 | No |
| 48 | Ppif | 5097 | 0.052 | 0.0822 | No |
| 49 | Irf1 | 5245 | 0.043 | 0.0711 | No |
| 50 | Eif5 | 5381 | 0.031 | 0.0605 | No |
| 51 | Dgat1 | 6002 | -0.002 | 0.0067 | No |
| 52 | Cyb5b | 6009 | -0.003 | 0.0063 | No |
| 53 | Tgfbrap1 | 6018 | -0.003 | 0.0057 | No |
| 54 | Eif2s3x | 6042 | -0.005 | 0.0039 | No |
| 55 | Fkbp4 | 6078 | -0.009 | 0.0012 | No |
| 56 | Plcl1 | 6100 | -0.010 | -0.0002 | No |
| 57 | Ago2 | 6107 | -0.010 | -0.0004 | No |
| 58 | Rfc4 | 6251 | -0.021 | -0.0120 | No |
| 59 | Ccnd3 | 6336 | -0.028 | -0.0183 | No |
| 60 | Cnp | 6524 | -0.041 | -0.0330 | No |
| 61 | Polr2h | 6726 | -0.055 | -0.0483 | No |
| 62 | Icam1 | 6741 | -0.058 | -0.0474 | No |
| 63 | Fen1 | 6880 | -0.068 | -0.0568 | No |
| 64 | Furin | 7019 | -0.078 | -0.0658 | No |
| 65 | Stard3 | 7211 | -0.093 | -0.0789 | No |
| 66 | Sqstm1 | 7254 | -0.096 | -0.0789 | No |
| 67 | Rpn1 | 7277 | -0.098 | -0.0771 | No |
| 68 | Btg1 | 7428 | -0.108 | -0.0860 | No |
| 69 | Polg2 | 7429 | -0.108 | -0.0819 | No |
| 70 | Rab27a | 7478 | -0.114 | -0.0818 | No |
| 71 | Nfkbia | 7696 | -0.129 | -0.0957 | No |
| 72 | Cdc5l | 7698 | -0.129 | -0.0909 | No |
| 73 | Maoa | 7912 | -0.147 | -0.1038 | No |
| 74 | Rxrb | 8000 | -0.153 | -0.1056 | No |
| 75 | Klhdc3 | 8066 | -0.158 | -0.1053 | No |
| 76 | Mgat1 | 8208 | -0.169 | -0.1111 | No |
| 77 | Cdc34 | 8273 | -0.174 | -0.1101 | No |
| 78 | Ppp1r2 | 8282 | -0.175 | -0.1042 | No |
| 79 | Creg1 | 8288 | -0.175 | -0.0979 | No |
| 80 | Mrpl23 | 8500 | -0.195 | -0.1089 | No |
| 81 | Alas1 | 8504 | -0.196 | -0.1017 | No |
| 82 | Hnrnpu | 8564 | -0.200 | -0.0992 | No |
| 83 | Tuba4a | 8609 | -0.204 | -0.0953 | No |
| 84 | Grpel1 | 8752 | -0.217 | -0.0994 | No |
| 85 | Urod | 8832 | -0.224 | -0.0978 | No |
| 86 | Cdk2 | 8994 | -0.235 | -0.1029 | No |
| 87 | Grina | 9005 | -0.237 | -0.0948 | No |
| 88 | Casp3 | 9175 | -0.245 | -0.1002 | No |
| 89 | Bsg | 9177 | -0.245 | -0.0910 | No |
| 90 | Cebpg | 9255 | -0.252 | -0.0881 | No |
| 91 | Mark2 | 9262 | -0.252 | -0.0790 | No |
| 92 | Dnajb1 | 9445 | -0.273 | -0.0845 | No |
| 93 | Btg3 | 9530 | -0.280 | -0.0812 | No |
| 94 | Ddx21 | 9840 | -0.315 | -0.0961 | No |
| 95 | Dlg4 | 10165 | -0.353 | -0.1109 | No |
| 96 | Hspa13 | 10396 | -0.387 | -0.1162 | No |
| 97 | Stip1 | 10513 | -0.405 | -0.1109 | No |
| 98 | Car2 | 10591 | -0.416 | -0.1018 | No |
| 99 | Tmbim6 | 10650 | -0.427 | -0.0906 | No |
| 100 | E2f5 | 10786 | -0.448 | -0.0854 | No |
| 101 | Prpf3 | 10904 | -0.470 | -0.0777 | No |
| 102 | Ap2s1 | 11052 | -0.501 | -0.0714 | No |
| 103 | Psmc3 | 11186 | -0.545 | -0.0623 | No |
| 104 | Bak1 | 11244 | -0.569 | -0.0457 | No |
| 105 | Cdo1 | 11341 | -0.606 | -0.0310 | No |
| 106 | Slc25a4 | 11348 | -0.611 | -0.0084 | No |
| 107 | Ctsl | 11546 | -0.817 | 0.0055 | No |