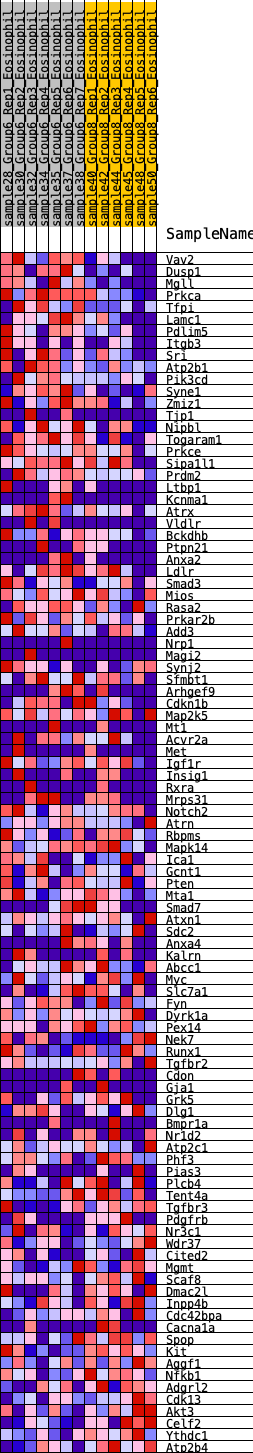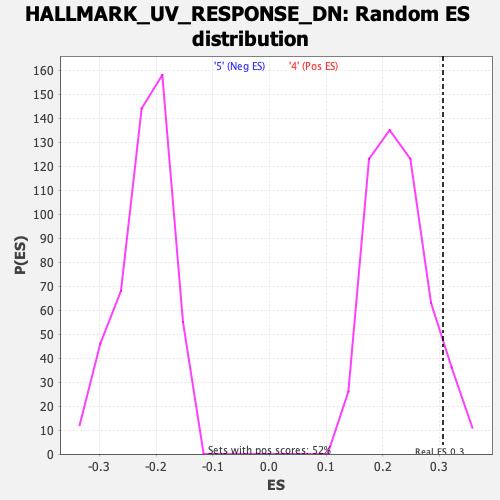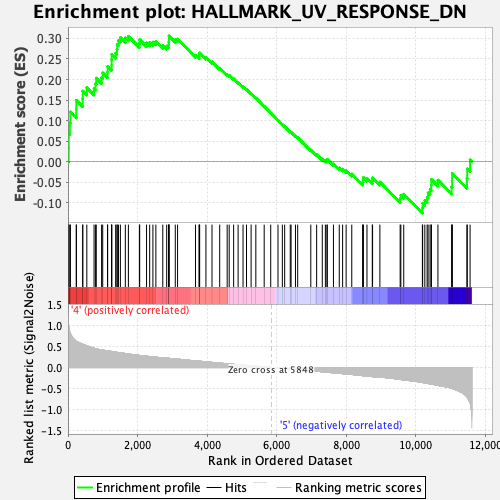
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group6_versus_Group8.Eosinophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Eosinophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | 0.3063579 |
| Normalized Enrichment Score (NES) | 1.3453132 |
| Nominal p-value | 0.08317215 |
| FDR q-value | 0.638995 |
| FWER p-Value | 0.596 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Vav2 | 19 | 1.010 | 0.0332 | Yes |
| 2 | Dusp1 | 20 | 1.002 | 0.0678 | Yes |
| 3 | Mgll | 53 | 0.843 | 0.0942 | Yes |
| 4 | Prkca | 69 | 0.814 | 0.1209 | Yes |
| 5 | Tfpi | 239 | 0.627 | 0.1279 | Yes |
| 6 | Lamc1 | 240 | 0.627 | 0.1496 | Yes |
| 7 | Pdlim5 | 422 | 0.557 | 0.1531 | Yes |
| 8 | Itgb3 | 425 | 0.555 | 0.1721 | Yes |
| 9 | Sri | 541 | 0.519 | 0.1800 | Yes |
| 10 | Atp2b1 | 749 | 0.460 | 0.1779 | Yes |
| 11 | Pik3cd | 793 | 0.450 | 0.1897 | Yes |
| 12 | Syne1 | 812 | 0.444 | 0.2035 | Yes |
| 13 | Zmiz1 | 966 | 0.419 | 0.2047 | Yes |
| 14 | Tjp1 | 998 | 0.415 | 0.2163 | Yes |
| 15 | Nipbl | 1137 | 0.399 | 0.2181 | Yes |
| 16 | Togaram1 | 1139 | 0.399 | 0.2318 | Yes |
| 17 | Prkce | 1253 | 0.384 | 0.2352 | Yes |
| 18 | Sipa1l1 | 1254 | 0.384 | 0.2485 | Yes |
| 19 | Prdm2 | 1258 | 0.383 | 0.2614 | Yes |
| 20 | Ltbp1 | 1370 | 0.368 | 0.2645 | Yes |
| 21 | Kcnma1 | 1407 | 0.362 | 0.2739 | Yes |
| 22 | Atrx | 1416 | 0.361 | 0.2856 | Yes |
| 23 | Vldlr | 1451 | 0.358 | 0.2950 | Yes |
| 24 | Bckdhb | 1506 | 0.350 | 0.3025 | Yes |
| 25 | Ptpn21 | 1648 | 0.334 | 0.3017 | Yes |
| 26 | Anxa2 | 1736 | 0.323 | 0.3053 | Yes |
| 27 | Ldlr | 2052 | 0.288 | 0.2879 | Yes |
| 28 | Smad3 | 2061 | 0.287 | 0.2972 | Yes |
| 29 | Mios | 2258 | 0.269 | 0.2894 | Yes |
| 30 | Rasa2 | 2348 | 0.262 | 0.2907 | Yes |
| 31 | Prkar2b | 2441 | 0.253 | 0.2914 | Yes |
| 32 | Add3 | 2526 | 0.247 | 0.2927 | Yes |
| 33 | Nrp1 | 2725 | 0.230 | 0.2834 | Yes |
| 34 | Magi2 | 2837 | 0.222 | 0.2815 | Yes |
| 35 | Synj2 | 2893 | 0.219 | 0.2842 | Yes |
| 36 | Sfmbt1 | 2898 | 0.219 | 0.2914 | Yes |
| 37 | Arhgef9 | 2901 | 0.218 | 0.2988 | Yes |
| 38 | Cdkn1b | 2902 | 0.218 | 0.3064 | Yes |
| 39 | Map2k5 | 3084 | 0.202 | 0.2976 | No |
| 40 | Mt1 | 3150 | 0.197 | 0.2988 | No |
| 41 | Acvr2a | 3666 | 0.158 | 0.2595 | No |
| 42 | Met | 3770 | 0.150 | 0.2557 | No |
| 43 | Igf1r | 3774 | 0.150 | 0.2607 | No |
| 44 | Insig1 | 3785 | 0.149 | 0.2649 | No |
| 45 | Rxra | 3965 | 0.136 | 0.2541 | No |
| 46 | Mrps31 | 4140 | 0.122 | 0.2432 | No |
| 47 | Notch2 | 4361 | 0.105 | 0.2277 | No |
| 48 | Atrn | 4576 | 0.089 | 0.2122 | No |
| 49 | Rbpms | 4634 | 0.086 | 0.2102 | No |
| 50 | Mapk14 | 4762 | 0.075 | 0.2018 | No |
| 51 | Ica1 | 4890 | 0.066 | 0.1930 | No |
| 52 | Gcnt1 | 5032 | 0.056 | 0.1827 | No |
| 53 | Pten | 5132 | 0.050 | 0.1758 | No |
| 54 | Mta1 | 5264 | 0.041 | 0.1659 | No |
| 55 | Smad7 | 5400 | 0.029 | 0.1551 | No |
| 56 | Atxn1 | 5640 | 0.013 | 0.1348 | No |
| 57 | Sdc2 | 5828 | 0.001 | 0.1186 | No |
| 58 | Anxa4 | 6037 | -0.005 | 0.1007 | No |
| 59 | Kalrn | 6162 | -0.014 | 0.0904 | No |
| 60 | Abcc1 | 6229 | -0.019 | 0.0854 | No |
| 61 | Myc | 6389 | -0.031 | 0.0726 | No |
| 62 | Slc7a1 | 6413 | -0.033 | 0.0718 | No |
| 63 | Fyn | 6539 | -0.042 | 0.0624 | No |
| 64 | Dyrk1a | 6603 | -0.047 | 0.0585 | No |
| 65 | Pex14 | 6980 | -0.075 | 0.0284 | No |
| 66 | Nek7 | 7148 | -0.089 | 0.0170 | No |
| 67 | Runx1 | 7310 | -0.100 | 0.0064 | No |
| 68 | Tgfbr2 | 7399 | -0.106 | 0.0025 | No |
| 69 | Cdon | 7444 | -0.110 | 0.0024 | No |
| 70 | Gja1 | 7454 | -0.111 | 0.0055 | No |
| 71 | Grk5 | 7631 | -0.124 | -0.0055 | No |
| 72 | Dlg1 | 7798 | -0.138 | -0.0152 | No |
| 73 | Bmpr1a | 7895 | -0.146 | -0.0185 | No |
| 74 | Nr1d2 | 7996 | -0.152 | -0.0219 | No |
| 75 | Atp2c1 | 8158 | -0.165 | -0.0302 | No |
| 76 | Phf3 | 8470 | -0.193 | -0.0505 | No |
| 77 | Pias3 | 8482 | -0.194 | -0.0448 | No |
| 78 | Plcb4 | 8491 | -0.195 | -0.0387 | No |
| 79 | Tent4a | 8595 | -0.203 | -0.0407 | No |
| 80 | Tgfbr3 | 8748 | -0.217 | -0.0464 | No |
| 81 | Pdgfrb | 8756 | -0.218 | -0.0395 | No |
| 82 | Nr3c1 | 8966 | -0.232 | -0.0496 | No |
| 83 | Wdr37 | 9549 | -0.283 | -0.0904 | No |
| 84 | Cited2 | 9569 | -0.285 | -0.0823 | No |
| 85 | Mgmt | 9652 | -0.295 | -0.0792 | No |
| 86 | Scaf8 | 10188 | -0.356 | -0.1134 | No |
| 87 | Dmac2l | 10196 | -0.357 | -0.1016 | No |
| 88 | Inpp4b | 10258 | -0.367 | -0.0943 | No |
| 89 | Cdc42bpa | 10324 | -0.376 | -0.0869 | No |
| 90 | Cacna1a | 10352 | -0.381 | -0.0761 | No |
| 91 | Spop | 10408 | -0.388 | -0.0675 | No |
| 92 | Kit | 10442 | -0.394 | -0.0567 | No |
| 93 | Aggf1 | 10447 | -0.395 | -0.0435 | No |
| 94 | Nfkb1 | 10635 | -0.426 | -0.0450 | No |
| 95 | Adgrl2 | 11029 | -0.494 | -0.0621 | No |
| 96 | Cdk13 | 11044 | -0.499 | -0.0461 | No |
| 97 | Akt3 | 11045 | -0.500 | -0.0288 | No |
| 98 | Celf2 | 11469 | -0.700 | -0.0414 | No |
| 99 | Ythdc1 | 11478 | -0.708 | -0.0176 | No |
| 100 | Atp2b4 | 11559 | -0.838 | 0.0043 | No |