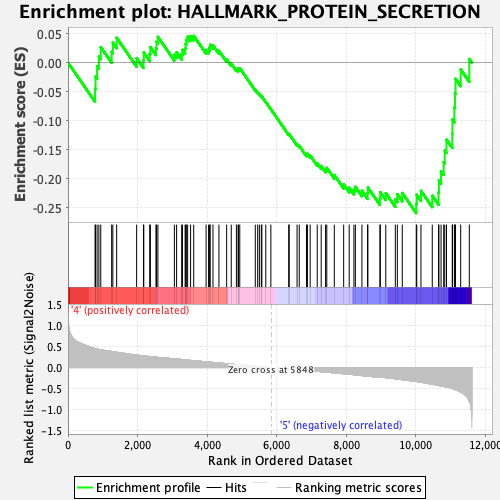
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group6_versus_Group8.Eosinophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Eosinophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.2592718 |
| Normalized Enrichment Score (NES) | -1.1819304 |
| Nominal p-value | 0.17391305 |
| FDR q-value | 0.46690315 |
| FWER p-Value | 0.842 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Atp7a | 778 | 0.454 | -0.0452 | No |
| 2 | Gosr2 | 792 | 0.450 | -0.0241 | No |
| 3 | Dnm1l | 837 | 0.439 | -0.0063 | No |
| 4 | Arfip1 | 888 | 0.432 | 0.0106 | No |
| 5 | Zw10 | 942 | 0.422 | 0.0268 | No |
| 6 | Tsg101 | 1255 | 0.384 | 0.0186 | No |
| 7 | Krt18 | 1289 | 0.378 | 0.0344 | No |
| 8 | Vps4b | 1399 | 0.363 | 0.0428 | No |
| 9 | Vps45 | 1973 | 0.296 | 0.0076 | No |
| 10 | Cltc | 2169 | 0.278 | 0.0044 | No |
| 11 | Rab14 | 2179 | 0.277 | 0.0173 | No |
| 12 | Stam | 2349 | 0.262 | 0.0155 | No |
| 13 | Sec24d | 2368 | 0.260 | 0.0267 | No |
| 14 | Atp6v1h | 2529 | 0.247 | 0.0250 | No |
| 15 | Lamp2 | 2546 | 0.245 | 0.0357 | No |
| 16 | Napa | 2584 | 0.241 | 0.0444 | No |
| 17 | Bnip3 | 3058 | 0.205 | 0.0134 | No |
| 18 | Stx12 | 3122 | 0.199 | 0.0178 | No |
| 19 | Atp1a1 | 3268 | 0.190 | 0.0145 | No |
| 20 | Snx2 | 3290 | 0.189 | 0.0220 | No |
| 21 | Yipf6 | 3367 | 0.183 | 0.0244 | No |
| 22 | Stx7 | 3382 | 0.181 | 0.0321 | No |
| 23 | Ap1g1 | 3402 | 0.179 | 0.0393 | No |
| 24 | Tpd52 | 3440 | 0.176 | 0.0447 | No |
| 25 | Ykt6 | 3524 | 0.168 | 0.0458 | No |
| 26 | Sec31a | 3614 | 0.162 | 0.0461 | No |
| 27 | Sgms1 | 3970 | 0.135 | 0.0219 | No |
| 28 | Stx16 | 4041 | 0.130 | 0.0223 | No |
| 29 | Ergic3 | 4070 | 0.128 | 0.0261 | No |
| 30 | Dop1a | 4088 | 0.126 | 0.0309 | No |
| 31 | Cope | 4168 | 0.120 | 0.0299 | No |
| 32 | Vamp4 | 4339 | 0.106 | 0.0204 | No |
| 33 | Ppt1 | 4561 | 0.091 | 0.0057 | No |
| 34 | Pam | 4693 | 0.081 | -0.0017 | No |
| 35 | Gbf1 | 4841 | 0.069 | -0.0111 | No |
| 36 | Ica1 | 4890 | 0.066 | -0.0120 | No |
| 37 | Arcn1 | 4900 | 0.065 | -0.0095 | No |
| 38 | Rps6ka3 | 4938 | 0.063 | -0.0096 | No |
| 39 | Arfgef2 | 5384 | 0.031 | -0.0467 | No |
| 40 | Gla | 5453 | 0.025 | -0.0514 | No |
| 41 | Napg | 5502 | 0.023 | -0.0544 | No |
| 42 | Mapk1 | 5558 | 0.019 | -0.0583 | No |
| 43 | Golga4 | 5573 | 0.018 | -0.0586 | No |
| 44 | Arfgap3 | 5686 | 0.010 | -0.0678 | No |
| 45 | Scamp3 | 5832 | 0.001 | -0.0804 | No |
| 46 | Rab5a | 6345 | -0.028 | -0.1234 | No |
| 47 | Tmx1 | 6359 | -0.029 | -0.1231 | No |
| 48 | Gnas | 6585 | -0.046 | -0.1404 | No |
| 49 | Vamp3 | 6648 | -0.050 | -0.1433 | No |
| 50 | Copb1 | 6856 | -0.067 | -0.1579 | No |
| 51 | Ap3b1 | 6883 | -0.069 | -0.1568 | No |
| 52 | Clcn3 | 6962 | -0.073 | -0.1600 | No |
| 53 | Rab2a | 7165 | -0.089 | -0.1731 | No |
| 54 | Cog2 | 7279 | -0.098 | -0.1781 | No |
| 55 | Cln5 | 7405 | -0.107 | -0.1837 | No |
| 56 | Dst | 7440 | -0.109 | -0.1812 | No |
| 57 | Adam10 | 7656 | -0.126 | -0.1937 | No |
| 58 | Arfgef1 | 7925 | -0.148 | -0.2096 | No |
| 59 | Copb2 | 8084 | -0.159 | -0.2155 | No |
| 60 | Scamp1 | 8220 | -0.170 | -0.2189 | No |
| 61 | Anp32e | 8261 | -0.173 | -0.2138 | No |
| 62 | Ap2m1 | 8450 | -0.191 | -0.2207 | No |
| 63 | Bet1 | 8612 | -0.204 | -0.2246 | No |
| 64 | Mon2 | 8621 | -0.205 | -0.2152 | No |
| 65 | Rer1 | 8967 | -0.232 | -0.2337 | No |
| 66 | Ap2b1 | 8979 | -0.234 | -0.2232 | No |
| 67 | Tom1l1 | 9136 | -0.242 | -0.2248 | No |
| 68 | Lman1 | 9410 | -0.269 | -0.2352 | No |
| 69 | Uso1 | 9468 | -0.275 | -0.2266 | No |
| 70 | Arf1 | 9610 | -0.291 | -0.2246 | No |
| 71 | Clta | 10011 | -0.332 | -0.2429 | Yes |
| 72 | Igf2r | 10026 | -0.334 | -0.2276 | Yes |
| 73 | Rab9 | 10147 | -0.350 | -0.2208 | Yes |
| 74 | Cd63 | 10472 | -0.400 | -0.2292 | Yes |
| 75 | Ap3s1 | 10653 | -0.428 | -0.2237 | Yes |
| 76 | M6pr | 10663 | -0.430 | -0.2033 | Yes |
| 77 | Sec22b | 10725 | -0.439 | -0.1870 | Yes |
| 78 | Sod1 | 10801 | -0.451 | -0.1713 | Yes |
| 79 | Abca1 | 10831 | -0.457 | -0.1513 | Yes |
| 80 | Ocrl | 10881 | -0.465 | -0.1326 | Yes |
| 81 | Galc | 11048 | -0.500 | -0.1224 | Yes |
| 82 | Ap2s1 | 11052 | -0.501 | -0.0980 | Yes |
| 83 | Kif1b | 11108 | -0.518 | -0.0772 | Yes |
| 84 | Ctsc | 11125 | -0.526 | -0.0527 | Yes |
| 85 | Snap23 | 11141 | -0.532 | -0.0278 | Yes |
| 86 | Tmed10 | 11289 | -0.586 | -0.0117 | Yes |
| 87 | Rab22a | 11537 | -0.800 | 0.0062 | Yes |