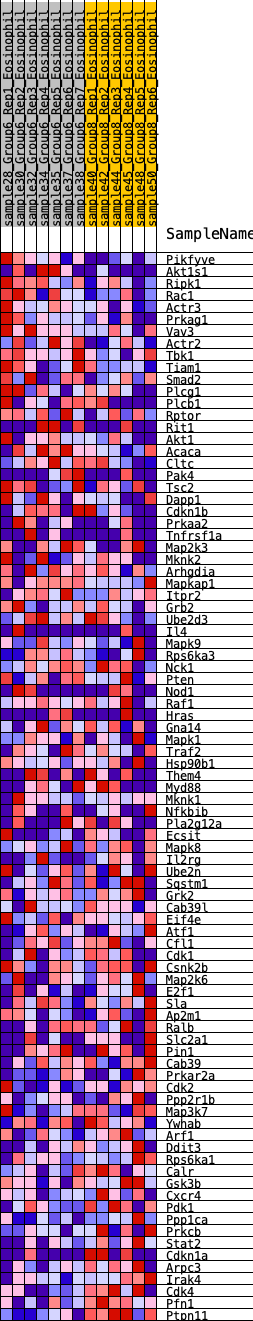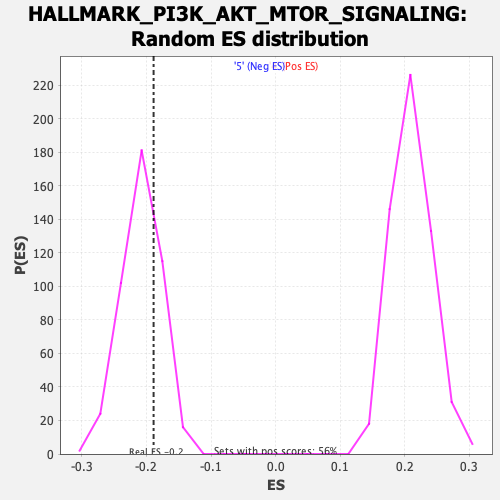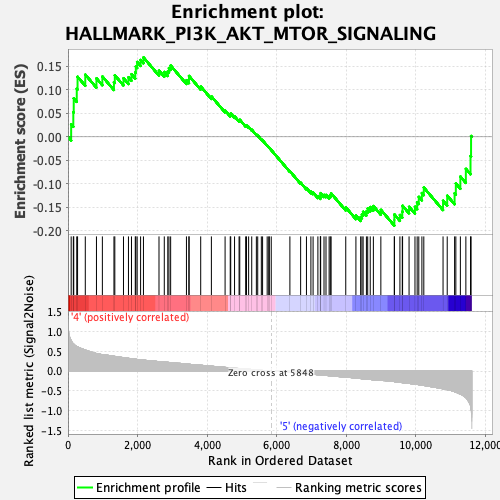
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group6_versus_Group8.Eosinophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Eosinophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.18893167 |
| Normalized Enrichment Score (NES) | -0.9052916 |
| Nominal p-value | 0.7295455 |
| FDR q-value | 0.8137923 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pikfyve | 91 | 0.773 | 0.0267 | No |
| 2 | Akt1s1 | 153 | 0.694 | 0.0525 | No |
| 3 | Ripk1 | 165 | 0.680 | 0.0819 | No |
| 4 | Rac1 | 252 | 0.622 | 0.1023 | No |
| 5 | Actr3 | 275 | 0.613 | 0.1278 | No |
| 6 | Prkag1 | 497 | 0.532 | 0.1324 | No |
| 7 | Vav3 | 818 | 0.443 | 0.1245 | No |
| 8 | Actr2 | 988 | 0.416 | 0.1284 | No |
| 9 | Tbk1 | 1321 | 0.375 | 0.1164 | No |
| 10 | Tiam1 | 1346 | 0.372 | 0.1309 | No |
| 11 | Smad2 | 1595 | 0.341 | 0.1247 | No |
| 12 | Plcg1 | 1738 | 0.323 | 0.1268 | No |
| 13 | Plcb1 | 1825 | 0.312 | 0.1333 | No |
| 14 | Rptor | 1934 | 0.301 | 0.1374 | No |
| 15 | Rit1 | 1950 | 0.299 | 0.1494 | No |
| 16 | Akt1 | 1988 | 0.294 | 0.1594 | No |
| 17 | Acaca | 2087 | 0.285 | 0.1636 | No |
| 18 | Cltc | 2169 | 0.278 | 0.1690 | No |
| 19 | Pak4 | 2616 | 0.238 | 0.1410 | No |
| 20 | Tsc2 | 2768 | 0.229 | 0.1381 | No |
| 21 | Dapp1 | 2869 | 0.221 | 0.1393 | No |
| 22 | Cdkn1b | 2902 | 0.218 | 0.1463 | No |
| 23 | Prkaa2 | 2951 | 0.214 | 0.1517 | No |
| 24 | Tnfrsf1a | 3405 | 0.179 | 0.1204 | No |
| 25 | Map2k3 | 3478 | 0.172 | 0.1219 | No |
| 26 | Mknk2 | 3479 | 0.172 | 0.1296 | No |
| 27 | Arhgdia | 3815 | 0.147 | 0.1071 | No |
| 28 | Mapkap1 | 4122 | 0.123 | 0.0861 | No |
| 29 | Itpr2 | 4516 | 0.094 | 0.0562 | No |
| 30 | Grb2 | 4664 | 0.083 | 0.0471 | No |
| 31 | Ube2d3 | 4678 | 0.082 | 0.0497 | No |
| 32 | Il4 | 4788 | 0.074 | 0.0435 | No |
| 33 | Mapk9 | 4918 | 0.064 | 0.0352 | No |
| 34 | Rps6ka3 | 4938 | 0.063 | 0.0364 | No |
| 35 | Nck1 | 5108 | 0.051 | 0.0240 | No |
| 36 | Pten | 5132 | 0.050 | 0.0242 | No |
| 37 | Nod1 | 5193 | 0.046 | 0.0211 | No |
| 38 | Raf1 | 5282 | 0.039 | 0.0152 | No |
| 39 | Hras | 5415 | 0.028 | 0.0050 | No |
| 40 | Gna14 | 5454 | 0.025 | 0.0028 | No |
| 41 | Mapk1 | 5558 | 0.019 | -0.0053 | No |
| 42 | Traf2 | 5594 | 0.017 | -0.0076 | No |
| 43 | Hsp90b1 | 5726 | 0.007 | -0.0186 | No |
| 44 | Them4 | 5771 | 0.005 | -0.0223 | No |
| 45 | Myd88 | 5788 | 0.004 | -0.0235 | No |
| 46 | Mknk1 | 5843 | 0.000 | -0.0281 | No |
| 47 | Nfkbib | 6379 | -0.031 | -0.0732 | No |
| 48 | Pla2g12a | 6688 | -0.052 | -0.0976 | No |
| 49 | Ecsit | 6855 | -0.067 | -0.1090 | No |
| 50 | Mapk8 | 6983 | -0.075 | -0.1166 | No |
| 51 | Il2rg | 7046 | -0.079 | -0.1185 | No |
| 52 | Ube2n | 7184 | -0.092 | -0.1263 | No |
| 53 | Sqstm1 | 7254 | -0.096 | -0.1279 | No |
| 54 | Grk2 | 7260 | -0.097 | -0.1240 | No |
| 55 | Cab39l | 7263 | -0.097 | -0.1199 | No |
| 56 | Eif4e | 7360 | -0.104 | -0.1236 | No |
| 57 | Atf1 | 7417 | -0.107 | -0.1236 | No |
| 58 | Cfl1 | 7507 | -0.115 | -0.1262 | No |
| 59 | Cdk1 | 7541 | -0.118 | -0.1238 | No |
| 60 | Csnk2b | 7564 | -0.120 | -0.1203 | No |
| 61 | Map2k6 | 7985 | -0.152 | -0.1500 | No |
| 62 | E2f1 | 8277 | -0.174 | -0.1674 | No |
| 63 | Sla | 8413 | -0.188 | -0.1707 | No |
| 64 | Ap2m1 | 8450 | -0.191 | -0.1653 | No |
| 65 | Ralb | 8484 | -0.194 | -0.1594 | No |
| 66 | Slc2a1 | 8579 | -0.201 | -0.1586 | No |
| 67 | Pin1 | 8616 | -0.204 | -0.1526 | No |
| 68 | Cab39 | 8689 | -0.211 | -0.1494 | No |
| 69 | Prkar2a | 8779 | -0.220 | -0.1473 | No |
| 70 | Cdk2 | 8994 | -0.235 | -0.1553 | No |
| 71 | Ppp2r1b | 9382 | -0.265 | -0.1771 | Yes |
| 72 | Map3k7 | 9383 | -0.265 | -0.1652 | Yes |
| 73 | Ywhab | 9542 | -0.282 | -0.1663 | Yes |
| 74 | Arf1 | 9610 | -0.291 | -0.1591 | Yes |
| 75 | Ddit3 | 9619 | -0.292 | -0.1467 | Yes |
| 76 | Rps6ka1 | 9805 | -0.311 | -0.1489 | Yes |
| 77 | Calr | 9975 | -0.330 | -0.1488 | Yes |
| 78 | Gsk3b | 10038 | -0.336 | -0.1391 | Yes |
| 79 | Cxcr4 | 10085 | -0.343 | -0.1278 | Yes |
| 80 | Pdk1 | 10174 | -0.354 | -0.1196 | Yes |
| 81 | Ppp1ca | 10229 | -0.363 | -0.1080 | Yes |
| 82 | Prkcb | 10782 | -0.447 | -0.1359 | Yes |
| 83 | Stat2 | 10902 | -0.469 | -0.1252 | Yes |
| 84 | Cdkn1a | 11114 | -0.522 | -0.1202 | Yes |
| 85 | Arpc3 | 11150 | -0.534 | -0.0993 | Yes |
| 86 | Irak4 | 11280 | -0.583 | -0.0845 | Yes |
| 87 | Cdk4 | 11438 | -0.670 | -0.0681 | Yes |
| 88 | Pfn1 | 11572 | -0.869 | -0.0408 | Yes |
| 89 | Ptpn11 | 11588 | -0.981 | 0.0018 | Yes |