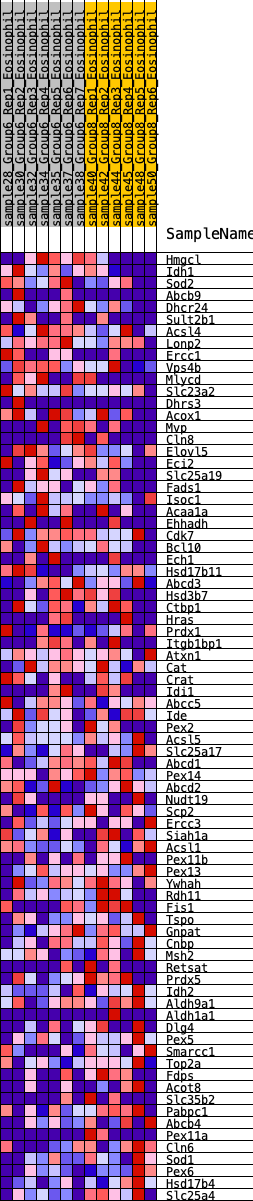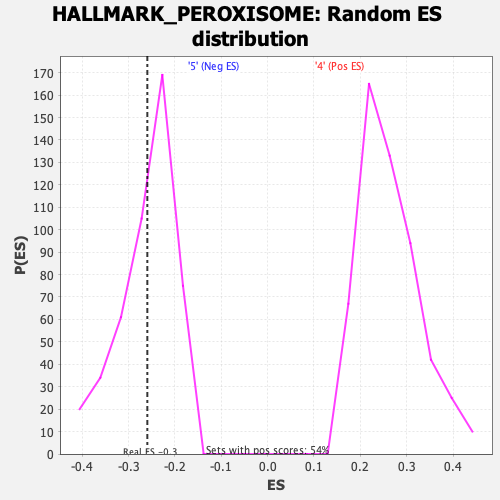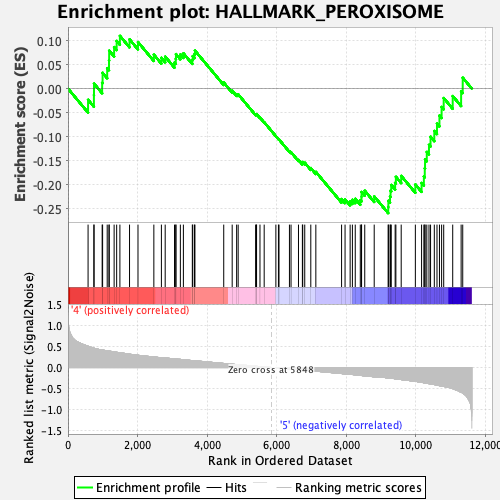
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group6_versus_Group8.Eosinophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Eosinophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.25956088 |
| Normalized Enrichment Score (NES) | -1.0030732 |
| Nominal p-value | 0.39439654 |
| FDR q-value | 0.758428 |
| FWER p-Value | 0.983 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hmgcl | 577 | 0.508 | -0.0234 | No |
| 2 | Idh1 | 745 | 0.462 | -0.0136 | No |
| 3 | Sod2 | 748 | 0.460 | 0.0104 | No |
| 4 | Abcb9 | 982 | 0.416 | 0.0121 | No |
| 5 | Dhcr24 | 995 | 0.415 | 0.0328 | No |
| 6 | Sult2b1 | 1127 | 0.401 | 0.0425 | No |
| 7 | Acsl4 | 1179 | 0.394 | 0.0588 | No |
| 8 | Lonp2 | 1184 | 0.393 | 0.0791 | No |
| 9 | Ercc1 | 1326 | 0.373 | 0.0864 | No |
| 10 | Vps4b | 1399 | 0.363 | 0.0993 | No |
| 11 | Mlycd | 1492 | 0.353 | 0.1098 | No |
| 12 | Slc23a2 | 1769 | 0.318 | 0.1026 | No |
| 13 | Dhrs3 | 2012 | 0.292 | 0.0969 | No |
| 14 | Acox1 | 2468 | 0.251 | 0.0707 | No |
| 15 | Mvp | 2685 | 0.232 | 0.0641 | No |
| 16 | Cln8 | 2794 | 0.226 | 0.0666 | No |
| 17 | Elovl5 | 3062 | 0.204 | 0.0542 | No |
| 18 | Eci2 | 3098 | 0.201 | 0.0617 | No |
| 19 | Slc25a19 | 3104 | 0.200 | 0.0718 | No |
| 20 | Fads1 | 3230 | 0.194 | 0.0712 | No |
| 21 | Isoc1 | 3317 | 0.186 | 0.0735 | No |
| 22 | Acaa1a | 3574 | 0.165 | 0.0599 | No |
| 23 | Ehhadh | 3586 | 0.164 | 0.0676 | No |
| 24 | Cdk7 | 3636 | 0.161 | 0.0718 | No |
| 25 | Bcl10 | 3649 | 0.159 | 0.0791 | No |
| 26 | Ech1 | 4477 | 0.097 | 0.0125 | No |
| 27 | Hsd17b11 | 4720 | 0.079 | -0.0043 | No |
| 28 | Abcd3 | 4848 | 0.069 | -0.0117 | No |
| 29 | Hsd3b7 | 4892 | 0.066 | -0.0120 | No |
| 30 | Ctbp1 | 5394 | 0.030 | -0.0539 | No |
| 31 | Hras | 5415 | 0.028 | -0.0542 | No |
| 32 | Prdx1 | 5421 | 0.028 | -0.0531 | No |
| 33 | Itgb1bp1 | 5519 | 0.021 | -0.0604 | No |
| 34 | Atxn1 | 5640 | 0.013 | -0.0701 | No |
| 35 | Cat | 5976 | -0.000 | -0.0992 | No |
| 36 | Crat | 6050 | -0.006 | -0.1052 | No |
| 37 | Idi1 | 6066 | -0.007 | -0.1061 | No |
| 38 | Abcc5 | 6366 | -0.030 | -0.1305 | No |
| 39 | Ide | 6412 | -0.033 | -0.1326 | No |
| 40 | Pex2 | 6626 | -0.049 | -0.1486 | No |
| 41 | Acsl5 | 6738 | -0.057 | -0.1552 | No |
| 42 | Slc25a17 | 6748 | -0.058 | -0.1529 | No |
| 43 | Abcd1 | 6806 | -0.063 | -0.1546 | No |
| 44 | Pex14 | 6980 | -0.075 | -0.1656 | No |
| 45 | Abcd2 | 7125 | -0.086 | -0.1736 | No |
| 46 | Nudt19 | 7865 | -0.143 | -0.2301 | No |
| 47 | Scp2 | 7965 | -0.150 | -0.2308 | No |
| 48 | Ercc3 | 8111 | -0.161 | -0.2349 | No |
| 49 | Siah1a | 8176 | -0.166 | -0.2317 | No |
| 50 | Acsl1 | 8258 | -0.173 | -0.2297 | No |
| 51 | Pex11b | 8403 | -0.187 | -0.2324 | No |
| 52 | Pex13 | 8438 | -0.190 | -0.2253 | No |
| 53 | Ywhah | 8440 | -0.190 | -0.2154 | No |
| 54 | Rdh11 | 8531 | -0.198 | -0.2128 | No |
| 55 | Fis1 | 8803 | -0.222 | -0.2247 | No |
| 56 | Tspo | 9206 | -0.247 | -0.2466 | Yes |
| 57 | Gnpat | 9210 | -0.247 | -0.2339 | Yes |
| 58 | Cnbp | 9258 | -0.252 | -0.2247 | Yes |
| 59 | Msh2 | 9273 | -0.253 | -0.2126 | Yes |
| 60 | Retsat | 9293 | -0.256 | -0.2008 | Yes |
| 61 | Prdx5 | 9405 | -0.269 | -0.1963 | Yes |
| 62 | Idh2 | 9426 | -0.271 | -0.1838 | Yes |
| 63 | Aldh9a1 | 9579 | -0.287 | -0.1819 | Yes |
| 64 | Aldh1a1 | 9986 | -0.330 | -0.1998 | Yes |
| 65 | Dlg4 | 10165 | -0.353 | -0.1967 | Yes |
| 66 | Pex5 | 10231 | -0.363 | -0.1832 | Yes |
| 67 | Smarcc1 | 10257 | -0.366 | -0.1661 | Yes |
| 68 | Top2a | 10265 | -0.368 | -0.1474 | Yes |
| 69 | Fdps | 10315 | -0.375 | -0.1320 | Yes |
| 70 | Acot8 | 10378 | -0.386 | -0.1171 | Yes |
| 71 | Slc35b2 | 10423 | -0.391 | -0.1003 | Yes |
| 72 | Pabpc1 | 10531 | -0.406 | -0.0883 | Yes |
| 73 | Abcb4 | 10608 | -0.420 | -0.0728 | Yes |
| 74 | Pex11a | 10679 | -0.433 | -0.0562 | Yes |
| 75 | Cln6 | 10741 | -0.441 | -0.0383 | Yes |
| 76 | Sod1 | 10801 | -0.451 | -0.0197 | Yes |
| 77 | Pex6 | 11060 | -0.504 | -0.0156 | Yes |
| 78 | Hsd17b4 | 11302 | -0.589 | -0.0055 | Yes |
| 79 | Slc25a4 | 11348 | -0.611 | 0.0226 | Yes |