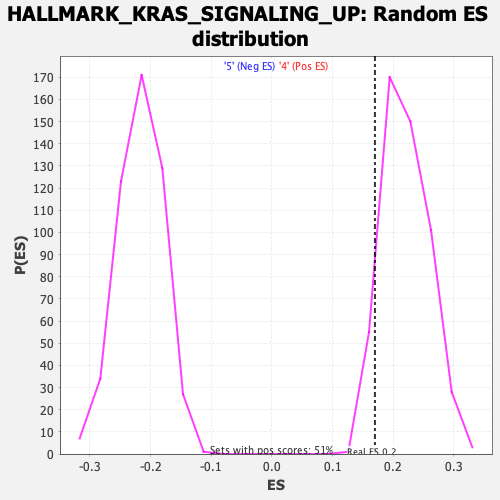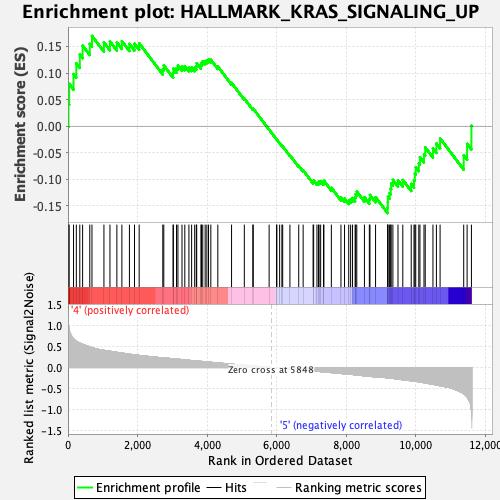
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group6_versus_Group8.Eosinophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Eosinophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | 0.16987166 |
| Normalized Enrichment Score (NES) | 0.7662949 |
| Nominal p-value | 0.9488189 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Spry2 | 14 | 1.023 | 0.0425 | Yes |
| 2 | Satb1 | 34 | 0.905 | 0.0796 | Yes |
| 3 | Crot | 159 | 0.687 | 0.0982 | Yes |
| 4 | Tfpi | 239 | 0.627 | 0.1182 | Yes |
| 5 | Pdcd1lg2 | 339 | 0.586 | 0.1347 | Yes |
| 6 | H2bc3 | 420 | 0.557 | 0.1516 | Yes |
| 7 | Atg10 | 626 | 0.493 | 0.1548 | Yes |
| 8 | Vwa5a | 689 | 0.477 | 0.1699 | Yes |
| 9 | Ank | 1033 | 0.411 | 0.1577 | No |
| 10 | Etv5 | 1206 | 0.389 | 0.1593 | No |
| 11 | Plvap | 1405 | 0.362 | 0.1576 | No |
| 12 | Galnt3 | 1549 | 0.346 | 0.1600 | No |
| 13 | Rbm4 | 1766 | 0.318 | 0.1549 | No |
| 14 | Cd37 | 1912 | 0.302 | 0.1552 | No |
| 15 | Ptbp2 | 2046 | 0.289 | 0.1560 | No |
| 16 | Nrp1 | 2725 | 0.230 | 0.1070 | No |
| 17 | Map4k1 | 2754 | 0.229 | 0.1143 | No |
| 18 | Btbd3 | 3021 | 0.208 | 0.1001 | No |
| 19 | Mtmr10 | 3029 | 0.207 | 0.1084 | No |
| 20 | Birc3 | 3120 | 0.199 | 0.1091 | No |
| 21 | Lat2 | 3160 | 0.196 | 0.1141 | No |
| 22 | Fbxo4 | 3277 | 0.189 | 0.1121 | No |
| 23 | Rabgap1l | 3358 | 0.184 | 0.1130 | No |
| 24 | Akt2 | 3476 | 0.173 | 0.1103 | No |
| 25 | Ikzf1 | 3554 | 0.166 | 0.1107 | No |
| 26 | Prelid3b | 3642 | 0.160 | 0.1100 | No |
| 27 | Pecam1 | 3689 | 0.157 | 0.1127 | No |
| 28 | Map3k1 | 3700 | 0.156 | 0.1185 | No |
| 29 | Ccser2 | 3822 | 0.147 | 0.1143 | No |
| 30 | Trib1 | 3838 | 0.146 | 0.1192 | No |
| 31 | Fcer1g | 3874 | 0.143 | 0.1223 | No |
| 32 | Map7 | 3943 | 0.138 | 0.1223 | No |
| 33 | Dock2 | 3992 | 0.134 | 0.1238 | No |
| 34 | Scg5 | 4039 | 0.130 | 0.1254 | No |
| 35 | Hsd11b1 | 4106 | 0.124 | 0.1250 | No |
| 36 | Itgb2 | 4307 | 0.109 | 0.1123 | No |
| 37 | Kcnn4 | 4702 | 0.080 | 0.0815 | No |
| 38 | Dusp6 | 5069 | 0.053 | 0.0520 | No |
| 39 | Ammecr1 | 5314 | 0.037 | 0.0324 | No |
| 40 | Zfp277 | 5325 | 0.036 | 0.0331 | No |
| 41 | Csf2ra | 5782 | 0.004 | -0.0064 | No |
| 42 | Ets1 | 5995 | -0.002 | -0.0247 | No |
| 43 | Laptm5 | 6005 | -0.002 | -0.0254 | No |
| 44 | Cbr4 | 6084 | -0.009 | -0.0318 | No |
| 45 | Tnfaip3 | 6149 | -0.013 | -0.0368 | No |
| 46 | Dnmbp | 6172 | -0.014 | -0.0381 | No |
| 47 | Fuca1 | 6380 | -0.031 | -0.0547 | No |
| 48 | Cmklr1 | 6633 | -0.049 | -0.0745 | No |
| 49 | Lcp1 | 6760 | -0.059 | -0.0829 | No |
| 50 | Il2rg | 7046 | -0.079 | -0.1043 | No |
| 51 | Cbl | 7062 | -0.081 | -0.1021 | No |
| 52 | Usp12 | 7157 | -0.089 | -0.1065 | No |
| 53 | Bpgm | 7200 | -0.093 | -0.1062 | No |
| 54 | Tnfrsf1b | 7218 | -0.094 | -0.1036 | No |
| 55 | Cab39l | 7263 | -0.097 | -0.1033 | No |
| 56 | Ero1a | 7345 | -0.103 | -0.1059 | No |
| 57 | Zfp639 | 7357 | -0.103 | -0.1025 | No |
| 58 | Tmem176a | 7571 | -0.121 | -0.1158 | No |
| 59 | Strn | 7847 | -0.142 | -0.1336 | No |
| 60 | Irf8 | 7950 | -0.149 | -0.1361 | No |
| 61 | Yrdc | 8070 | -0.158 | -0.1396 | No |
| 62 | Nin | 8125 | -0.162 | -0.1374 | No |
| 63 | Plaur | 8180 | -0.167 | -0.1349 | No |
| 64 | Psmb8 | 8254 | -0.172 | -0.1339 | No |
| 65 | Ppp1r15a | 8271 | -0.174 | -0.1279 | No |
| 66 | Gypc | 8302 | -0.176 | -0.1229 | No |
| 67 | Mmd | 8522 | -0.197 | -0.1335 | No |
| 68 | St6gal1 | 8663 | -0.209 | -0.1367 | No |
| 69 | Tspan13 | 8681 | -0.210 | -0.1292 | No |
| 70 | Angptl4 | 8842 | -0.225 | -0.1334 | No |
| 71 | Ptcd2 | 9189 | -0.246 | -0.1530 | No |
| 72 | F13a1 | 9194 | -0.246 | -0.1428 | No |
| 73 | Adgrl4 | 9198 | -0.246 | -0.1325 | No |
| 74 | Ccnd2 | 9245 | -0.251 | -0.1258 | No |
| 75 | Glrx | 9277 | -0.254 | -0.1176 | No |
| 76 | Eng | 9290 | -0.256 | -0.1077 | No |
| 77 | Avl9 | 9340 | -0.260 | -0.1008 | No |
| 78 | Evi5 | 9487 | -0.277 | -0.1017 | No |
| 79 | Adam8 | 9625 | -0.293 | -0.1010 | No |
| 80 | Cdadc1 | 9867 | -0.318 | -0.1084 | No |
| 81 | Itga2 | 9947 | -0.327 | -0.1013 | No |
| 82 | Adam17 | 9971 | -0.329 | -0.0892 | No |
| 83 | Il1rl2 | 9997 | -0.331 | -0.0772 | No |
| 84 | Cxcr4 | 10085 | -0.343 | -0.0701 | No |
| 85 | Tor1aip2 | 10118 | -0.346 | -0.0580 | No |
| 86 | Cbx8 | 10234 | -0.364 | -0.0525 | No |
| 87 | Ano1 | 10270 | -0.368 | -0.0398 | No |
| 88 | Tmem176b | 10492 | -0.404 | -0.0417 | No |
| 89 | Car2 | 10591 | -0.416 | -0.0324 | No |
| 90 | Jup | 10697 | -0.436 | -0.0229 | No |
| 91 | Wdr33 | 11376 | -0.628 | -0.0549 | No |
| 92 | Hdac9 | 11474 | -0.707 | -0.0331 | No |
| 93 | Sdccag8 | 11598 | -1.046 | 0.0010 | No |