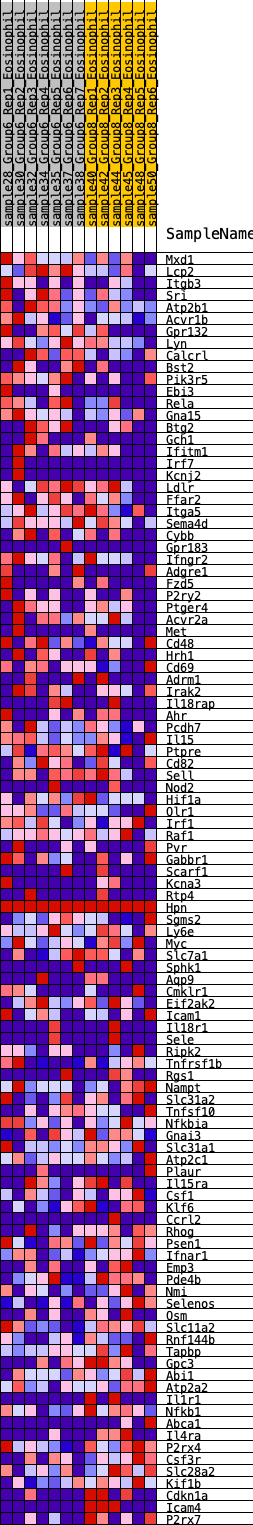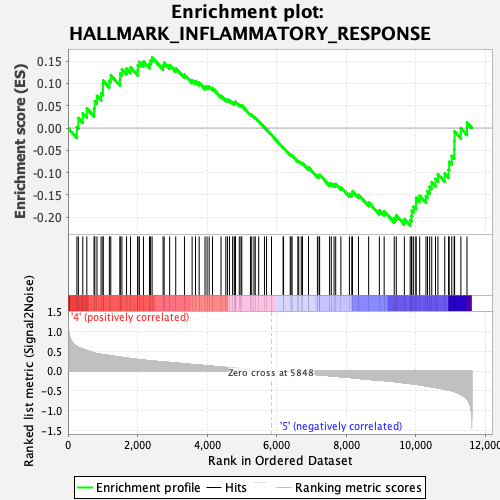
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group6_versus_Group8.Eosinophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Eosinophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | -0.2186318 |
| Normalized Enrichment Score (NES) | -0.96205354 |
| Nominal p-value | 0.5627615 |
| FDR q-value | 0.785221 |
| FWER p-Value | 0.993 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mxd1 | 256 | 0.621 | 0.0022 | No |
| 2 | Lcp2 | 298 | 0.601 | 0.0223 | No |
| 3 | Itgb3 | 425 | 0.555 | 0.0332 | No |
| 4 | Sri | 541 | 0.519 | 0.0437 | No |
| 5 | Atp2b1 | 749 | 0.460 | 0.0438 | No |
| 6 | Acvr1b | 771 | 0.456 | 0.0599 | No |
| 7 | Gpr132 | 833 | 0.441 | 0.0720 | No |
| 8 | Lyn | 951 | 0.421 | 0.0784 | No |
| 9 | Calcrl | 1008 | 0.414 | 0.0899 | No |
| 10 | Bst2 | 1009 | 0.414 | 0.1062 | No |
| 11 | Pik3r5 | 1190 | 0.391 | 0.1059 | No |
| 12 | Ebi3 | 1230 | 0.386 | 0.1177 | No |
| 13 | Rela | 1495 | 0.352 | 0.1087 | No |
| 14 | Gna15 | 1500 | 0.351 | 0.1222 | No |
| 15 | Btg2 | 1550 | 0.345 | 0.1315 | No |
| 16 | Gch1 | 1683 | 0.329 | 0.1330 | No |
| 17 | Ifitm1 | 1798 | 0.315 | 0.1355 | No |
| 18 | Irf7 | 2004 | 0.292 | 0.1292 | No |
| 19 | Kcnj2 | 2007 | 0.292 | 0.1405 | No |
| 20 | Ldlr | 2052 | 0.288 | 0.1481 | No |
| 21 | Ffar2 | 2170 | 0.278 | 0.1488 | No |
| 22 | Itga5 | 2342 | 0.262 | 0.1443 | No |
| 23 | Sema4d | 2375 | 0.259 | 0.1517 | No |
| 24 | Cybb | 2418 | 0.255 | 0.1582 | No |
| 25 | Gpr183 | 2733 | 0.230 | 0.1399 | No |
| 26 | Ifngr2 | 2764 | 0.229 | 0.1463 | No |
| 27 | Adgre1 | 2922 | 0.217 | 0.1412 | No |
| 28 | Fzd5 | 3097 | 0.201 | 0.1340 | No |
| 29 | P2ry2 | 3348 | 0.184 | 0.1195 | No |
| 30 | Ptger4 | 3568 | 0.165 | 0.1070 | No |
| 31 | Acvr2a | 3666 | 0.158 | 0.1048 | No |
| 32 | Met | 3770 | 0.150 | 0.1018 | No |
| 33 | Cd48 | 3939 | 0.138 | 0.0926 | No |
| 34 | Hrh1 | 3998 | 0.133 | 0.0928 | No |
| 35 | Cd69 | 4058 | 0.129 | 0.0928 | No |
| 36 | Adrm1 | 4154 | 0.121 | 0.0893 | No |
| 37 | Irak2 | 4399 | 0.102 | 0.0721 | No |
| 38 | Il18rap | 4537 | 0.092 | 0.0638 | No |
| 39 | Ahr | 4583 | 0.089 | 0.0634 | No |
| 40 | Pcdh7 | 4642 | 0.085 | 0.0617 | No |
| 41 | Il15 | 4732 | 0.078 | 0.0570 | No |
| 42 | Ptpre | 4780 | 0.074 | 0.0559 | No |
| 43 | Cd82 | 4803 | 0.072 | 0.0568 | No |
| 44 | Sell | 4814 | 0.072 | 0.0588 | No |
| 45 | Nod2 | 4924 | 0.064 | 0.0518 | No |
| 46 | Hif1a | 4972 | 0.060 | 0.0501 | No |
| 47 | Olr1 | 4996 | 0.059 | 0.0504 | No |
| 48 | Irf1 | 5245 | 0.043 | 0.0305 | No |
| 49 | Raf1 | 5282 | 0.039 | 0.0289 | No |
| 50 | Pvr | 5346 | 0.034 | 0.0248 | No |
| 51 | Gabbr1 | 5383 | 0.031 | 0.0229 | No |
| 52 | Scarf1 | 5482 | 0.024 | 0.0153 | No |
| 53 | Kcna3 | 5648 | 0.012 | 0.0015 | No |
| 54 | Rtp4 | 5708 | 0.008 | -0.0034 | No |
| 55 | Hpn | 5850 | 0.000 | -0.0156 | No |
| 56 | Sgms2 | 6184 | -0.015 | -0.0440 | No |
| 57 | Ly6e | 6197 | -0.017 | -0.0443 | No |
| 58 | Myc | 6389 | -0.031 | -0.0597 | No |
| 59 | Slc7a1 | 6413 | -0.033 | -0.0604 | No |
| 60 | Sphk1 | 6442 | -0.035 | -0.0615 | No |
| 61 | Aqp9 | 6608 | -0.047 | -0.0740 | No |
| 62 | Cmklr1 | 6633 | -0.049 | -0.0741 | No |
| 63 | Eif2ak2 | 6705 | -0.054 | -0.0782 | No |
| 64 | Icam1 | 6741 | -0.058 | -0.0789 | No |
| 65 | Il18r1 | 6914 | -0.071 | -0.0911 | No |
| 66 | Sele | 6915 | -0.071 | -0.0883 | No |
| 67 | Ripk2 | 7170 | -0.090 | -0.1068 | No |
| 68 | Tnfrsf1b | 7218 | -0.094 | -0.1072 | No |
| 69 | Rgs1 | 7234 | -0.095 | -0.1048 | No |
| 70 | Nampt | 7520 | -0.116 | -0.1250 | No |
| 71 | Slc31a2 | 7569 | -0.121 | -0.1244 | No |
| 72 | Tnfsf10 | 7648 | -0.125 | -0.1262 | No |
| 73 | Nfkbia | 7696 | -0.129 | -0.1252 | No |
| 74 | Gnai3 | 7845 | -0.142 | -0.1325 | No |
| 75 | Slc31a1 | 8091 | -0.159 | -0.1475 | No |
| 76 | Atp2c1 | 8158 | -0.165 | -0.1467 | No |
| 77 | Plaur | 8180 | -0.167 | -0.1420 | No |
| 78 | Il15ra | 8351 | -0.181 | -0.1496 | No |
| 79 | Csf1 | 8643 | -0.208 | -0.1667 | No |
| 80 | Klf6 | 8951 | -0.231 | -0.1843 | No |
| 81 | Ccrl2 | 9089 | -0.239 | -0.1868 | No |
| 82 | Rhog | 9379 | -0.264 | -0.2015 | No |
| 83 | Psen1 | 9437 | -0.272 | -0.1958 | No |
| 84 | Ifnar1 | 9670 | -0.297 | -0.2042 | No |
| 85 | Emp3 | 9837 | -0.315 | -0.2062 | Yes |
| 86 | Pde4b | 9875 | -0.318 | -0.1969 | Yes |
| 87 | Nmi | 9886 | -0.319 | -0.1852 | Yes |
| 88 | Selenos | 9936 | -0.326 | -0.1766 | Yes |
| 89 | Osm | 10003 | -0.332 | -0.1693 | Yes |
| 90 | Slc11a2 | 10005 | -0.332 | -0.1563 | Yes |
| 91 | Rnf144b | 10111 | -0.345 | -0.1518 | Yes |
| 92 | Tapbp | 10291 | -0.372 | -0.1527 | Yes |
| 93 | Gpc3 | 10335 | -0.378 | -0.1416 | Yes |
| 94 | Abi1 | 10397 | -0.387 | -0.1317 | Yes |
| 95 | Atp2a2 | 10457 | -0.398 | -0.1211 | Yes |
| 96 | Il1r1 | 10565 | -0.413 | -0.1141 | Yes |
| 97 | Nfkb1 | 10635 | -0.426 | -0.1033 | Yes |
| 98 | Abca1 | 10831 | -0.457 | -0.1023 | Yes |
| 99 | Il4ra | 10941 | -0.475 | -0.0930 | Yes |
| 100 | P2rx4 | 10959 | -0.478 | -0.0757 | Yes |
| 101 | Csf3r | 11033 | -0.495 | -0.0625 | Yes |
| 102 | Slc28a2 | 11104 | -0.517 | -0.0482 | Yes |
| 103 | Kif1b | 11108 | -0.518 | -0.0281 | Yes |
| 104 | Cdkn1a | 11114 | -0.522 | -0.0080 | Yes |
| 105 | Icam4 | 11295 | -0.589 | -0.0004 | Yes |
| 106 | P2rx7 | 11470 | -0.701 | 0.0121 | Yes |