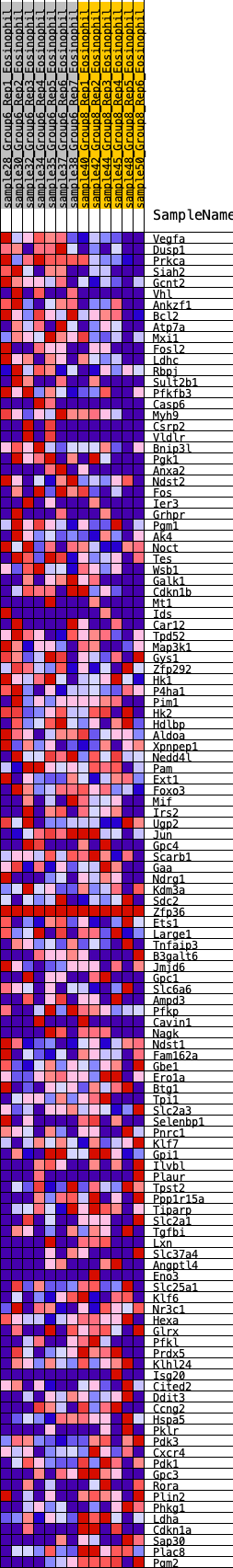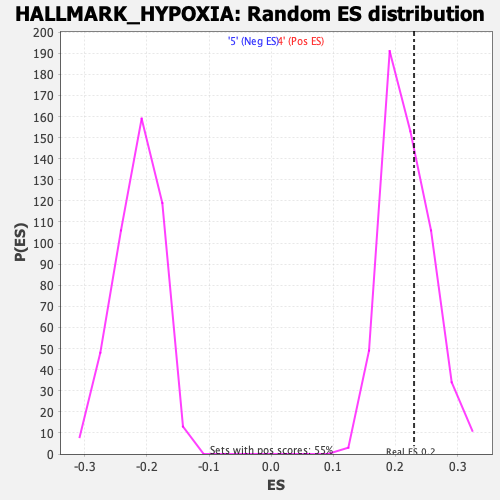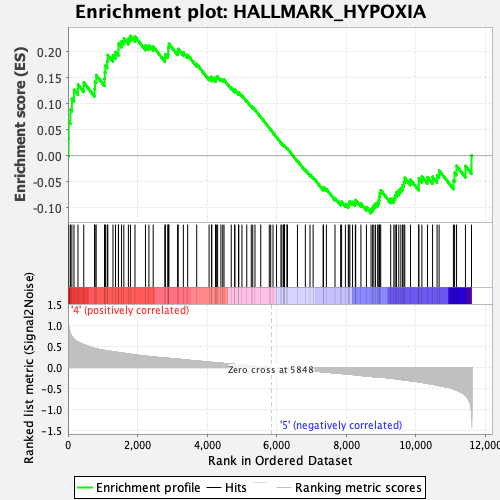
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group6_versus_Group8.Eosinophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Eosinophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_HYPOXIA |
| Enrichment Score (ES) | 0.22996144 |
| Normalized Enrichment Score (NES) | 1.0492331 |
| Nominal p-value | 0.34917733 |
| FDR q-value | 0.80819416 |
| FWER p-Value | 0.975 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Vegfa | 16 | 1.020 | 0.0325 | Yes |
| 2 | Dusp1 | 20 | 1.002 | 0.0655 | Yes |
| 3 | Prkca | 69 | 0.814 | 0.0883 | Yes |
| 4 | Siah2 | 111 | 0.742 | 0.1094 | Yes |
| 5 | Gcnt2 | 172 | 0.675 | 0.1266 | Yes |
| 6 | Vhl | 287 | 0.606 | 0.1368 | Yes |
| 7 | Ankzf1 | 453 | 0.545 | 0.1406 | Yes |
| 8 | Bcl2 | 762 | 0.457 | 0.1290 | Yes |
| 9 | Atp7a | 778 | 0.454 | 0.1427 | Yes |
| 10 | Mxi1 | 810 | 0.445 | 0.1548 | Yes |
| 11 | Fosl2 | 1050 | 0.409 | 0.1476 | Yes |
| 12 | Ldhc | 1058 | 0.409 | 0.1605 | Yes |
| 13 | Rbpj | 1074 | 0.407 | 0.1727 | Yes |
| 14 | Sult2b1 | 1127 | 0.401 | 0.1815 | Yes |
| 15 | Pfkfb3 | 1144 | 0.399 | 0.1934 | Yes |
| 16 | Casp6 | 1293 | 0.378 | 0.1931 | Yes |
| 17 | Myh9 | 1366 | 0.368 | 0.1990 | Yes |
| 18 | Csrp2 | 1450 | 0.358 | 0.2037 | Yes |
| 19 | Vldlr | 1451 | 0.358 | 0.2156 | Yes |
| 20 | Bnip3l | 1539 | 0.346 | 0.2195 | Yes |
| 21 | Pgk1 | 1605 | 0.340 | 0.2251 | Yes |
| 22 | Anxa2 | 1736 | 0.323 | 0.2245 | Yes |
| 23 | Ndst2 | 1795 | 0.315 | 0.2300 | Yes |
| 24 | Fos | 1926 | 0.301 | 0.2286 | No |
| 25 | Ier3 | 2227 | 0.273 | 0.2116 | No |
| 26 | Grhpr | 2326 | 0.263 | 0.2118 | No |
| 27 | Pgm1 | 2449 | 0.252 | 0.2096 | No |
| 28 | Ak4 | 2784 | 0.227 | 0.1880 | No |
| 29 | Noct | 2800 | 0.226 | 0.1942 | No |
| 30 | Tes | 2871 | 0.221 | 0.1955 | No |
| 31 | Wsb1 | 2882 | 0.220 | 0.2019 | No |
| 32 | Galk1 | 2883 | 0.220 | 0.2092 | No |
| 33 | Cdkn1b | 2902 | 0.218 | 0.2149 | No |
| 34 | Mt1 | 3150 | 0.197 | 0.2000 | No |
| 35 | Ids | 3168 | 0.195 | 0.2050 | No |
| 36 | Car12 | 3316 | 0.186 | 0.1984 | No |
| 37 | Tpd52 | 3440 | 0.176 | 0.1935 | No |
| 38 | Map3k1 | 3700 | 0.156 | 0.1761 | No |
| 39 | Gys1 | 4056 | 0.129 | 0.1495 | No |
| 40 | Zfp292 | 4130 | 0.123 | 0.1472 | No |
| 41 | Hk1 | 4131 | 0.123 | 0.1513 | No |
| 42 | P4ha1 | 4242 | 0.114 | 0.1455 | No |
| 43 | Pim1 | 4244 | 0.114 | 0.1492 | No |
| 44 | Hk2 | 4265 | 0.111 | 0.1512 | No |
| 45 | Hdlbp | 4301 | 0.109 | 0.1518 | No |
| 46 | Aldoa | 4396 | 0.102 | 0.1470 | No |
| 47 | Xpnpep1 | 4449 | 0.098 | 0.1457 | No |
| 48 | Nedd4l | 4485 | 0.096 | 0.1459 | No |
| 49 | Pam | 4693 | 0.081 | 0.1305 | No |
| 50 | Ext1 | 4794 | 0.073 | 0.1243 | No |
| 51 | Foxo3 | 4795 | 0.073 | 0.1267 | No |
| 52 | Mif | 4902 | 0.065 | 0.1196 | No |
| 53 | Irs2 | 4910 | 0.065 | 0.1212 | No |
| 54 | Ugp2 | 5001 | 0.058 | 0.1153 | No |
| 55 | Jun | 5139 | 0.050 | 0.1050 | No |
| 56 | Gpc4 | 5269 | 0.040 | 0.0951 | No |
| 57 | Scarb1 | 5311 | 0.037 | 0.0928 | No |
| 58 | Gaa | 5375 | 0.032 | 0.0883 | No |
| 59 | Ndrg1 | 5541 | 0.020 | 0.0746 | No |
| 60 | Kdm3a | 5787 | 0.004 | 0.0534 | No |
| 61 | Sdc2 | 5828 | 0.001 | 0.0500 | No |
| 62 | Zfp36 | 5894 | 0.000 | 0.0443 | No |
| 63 | Ets1 | 5995 | -0.002 | 0.0357 | No |
| 64 | Large1 | 6122 | -0.011 | 0.0251 | No |
| 65 | Tnfaip3 | 6149 | -0.013 | 0.0232 | No |
| 66 | B3galt6 | 6200 | -0.017 | 0.0195 | No |
| 67 | Jmjd6 | 6201 | -0.017 | 0.0200 | No |
| 68 | Gpc1 | 6222 | -0.019 | 0.0189 | No |
| 69 | Slc6a6 | 6225 | -0.019 | 0.0194 | No |
| 70 | Ampd3 | 6293 | -0.024 | 0.0143 | No |
| 71 | Pfkp | 6310 | -0.026 | 0.0138 | No |
| 72 | Cavin1 | 6596 | -0.046 | -0.0095 | No |
| 73 | Nagk | 6823 | -0.065 | -0.0270 | No |
| 74 | Ndst1 | 6957 | -0.073 | -0.0362 | No |
| 75 | Fam162a | 7047 | -0.079 | -0.0413 | No |
| 76 | Gbe1 | 7334 | -0.102 | -0.0628 | No |
| 77 | Ero1a | 7345 | -0.103 | -0.0602 | No |
| 78 | Btg1 | 7428 | -0.108 | -0.0638 | No |
| 79 | Tpi1 | 7676 | -0.127 | -0.0810 | No |
| 80 | Slc2a3 | 7838 | -0.141 | -0.0904 | No |
| 81 | Selenbp1 | 7866 | -0.143 | -0.0879 | No |
| 82 | Pnrc1 | 7975 | -0.151 | -0.0923 | No |
| 83 | Klf7 | 8058 | -0.157 | -0.0943 | No |
| 84 | Gpi1 | 8076 | -0.158 | -0.0905 | No |
| 85 | Ilvbl | 8103 | -0.160 | -0.0874 | No |
| 86 | Plaur | 8180 | -0.167 | -0.0885 | No |
| 87 | Tpst2 | 8260 | -0.173 | -0.0896 | No |
| 88 | Ppp1r15a | 8271 | -0.174 | -0.0847 | No |
| 89 | Tiparp | 8421 | -0.189 | -0.0914 | No |
| 90 | Slc2a1 | 8579 | -0.201 | -0.0984 | No |
| 91 | Tgfbi | 8712 | -0.213 | -0.1028 | No |
| 92 | Lxn | 8758 | -0.218 | -0.0995 | No |
| 93 | Slc37a4 | 8793 | -0.221 | -0.0951 | No |
| 94 | Angptl4 | 8842 | -0.225 | -0.0918 | No |
| 95 | Eno3 | 8903 | -0.228 | -0.0895 | No |
| 96 | Slc25a1 | 8937 | -0.230 | -0.0847 | No |
| 97 | Klf6 | 8951 | -0.231 | -0.0782 | No |
| 98 | Nr3c1 | 8966 | -0.232 | -0.0717 | No |
| 99 | Hexa | 8990 | -0.235 | -0.0659 | No |
| 100 | Glrx | 9277 | -0.254 | -0.0823 | No |
| 101 | Pfkl | 9368 | -0.263 | -0.0814 | No |
| 102 | Prdx5 | 9405 | -0.269 | -0.0756 | No |
| 103 | Klhl24 | 9443 | -0.273 | -0.0698 | No |
| 104 | Isg20 | 9510 | -0.279 | -0.0663 | No |
| 105 | Cited2 | 9569 | -0.285 | -0.0619 | No |
| 106 | Ddit3 | 9619 | -0.292 | -0.0564 | No |
| 107 | Ccng2 | 9657 | -0.295 | -0.0498 | No |
| 108 | Hspa5 | 9682 | -0.299 | -0.0420 | No |
| 109 | Pklr | 9846 | -0.316 | -0.0457 | No |
| 110 | Pdk3 | 10083 | -0.343 | -0.0549 | No |
| 111 | Cxcr4 | 10085 | -0.343 | -0.0435 | No |
| 112 | Pdk1 | 10174 | -0.354 | -0.0394 | No |
| 113 | Gpc3 | 10335 | -0.378 | -0.0408 | No |
| 114 | Rora | 10481 | -0.402 | -0.0401 | No |
| 115 | Plin2 | 10611 | -0.420 | -0.0374 | No |
| 116 | Phkg1 | 10669 | -0.432 | -0.0280 | No |
| 117 | Ldha | 11081 | -0.510 | -0.0468 | No |
| 118 | Cdkn1a | 11114 | -0.522 | -0.0322 | No |
| 119 | Sap30 | 11170 | -0.539 | -0.0191 | No |
| 120 | Plac8 | 11426 | -0.662 | -0.0193 | No |
| 121 | Pgm2 | 11600 | -1.059 | 0.0008 | No |