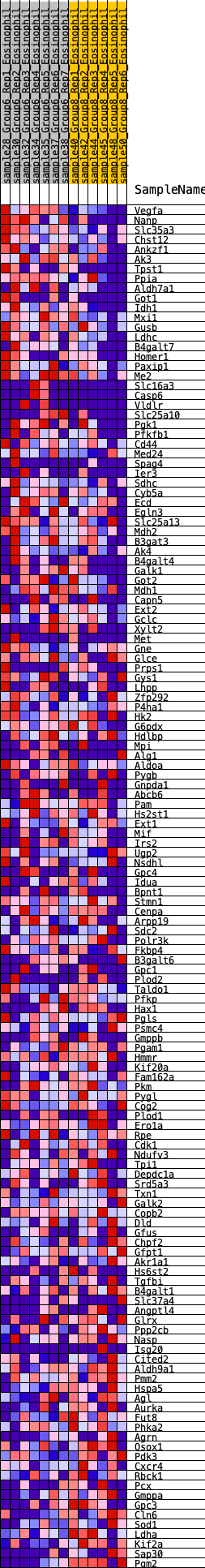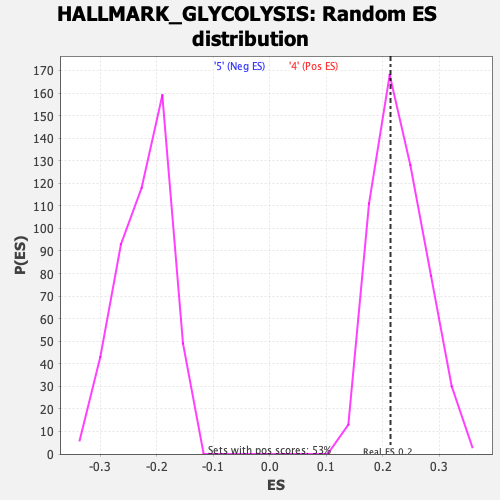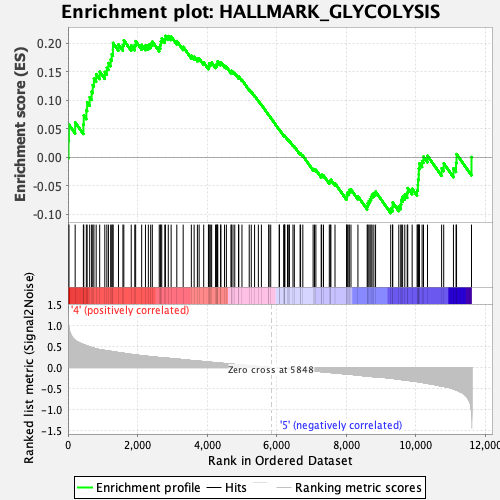
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group6_versus_Group8.Eosinophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Eosinophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_GLYCOLYSIS |
| Enrichment Score (ES) | 0.21335106 |
| Normalized Enrichment Score (NES) | 0.93187547 |
| Nominal p-value | 0.5864662 |
| FDR q-value | 0.87814015 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Vegfa | 16 | 1.020 | 0.0296 | Yes |
| 2 | Nanp | 27 | 0.945 | 0.0575 | Yes |
| 3 | Slc35a3 | 206 | 0.645 | 0.0616 | Yes |
| 4 | Chst12 | 442 | 0.550 | 0.0578 | Yes |
| 5 | Ankzf1 | 453 | 0.545 | 0.0736 | Yes |
| 6 | Ak3 | 527 | 0.523 | 0.0831 | Yes |
| 7 | Tpst1 | 550 | 0.518 | 0.0969 | Yes |
| 8 | Ppia | 623 | 0.495 | 0.1057 | Yes |
| 9 | Aldh7a1 | 679 | 0.480 | 0.1155 | Yes |
| 10 | Got1 | 712 | 0.471 | 0.1271 | Yes |
| 11 | Idh1 | 745 | 0.462 | 0.1383 | Yes |
| 12 | Mxi1 | 810 | 0.445 | 0.1463 | Yes |
| 13 | Gusb | 911 | 0.428 | 0.1506 | Yes |
| 14 | Ldhc | 1058 | 0.409 | 0.1503 | Yes |
| 15 | B4galt7 | 1117 | 0.402 | 0.1575 | Yes |
| 16 | Homer1 | 1162 | 0.397 | 0.1657 | Yes |
| 17 | Paxip1 | 1227 | 0.386 | 0.1719 | Yes |
| 18 | Me2 | 1257 | 0.383 | 0.1810 | Yes |
| 19 | Slc16a3 | 1291 | 0.378 | 0.1896 | Yes |
| 20 | Casp6 | 1293 | 0.378 | 0.2010 | Yes |
| 21 | Vldlr | 1451 | 0.358 | 0.1982 | Yes |
| 22 | Slc25a10 | 1579 | 0.342 | 0.1976 | Yes |
| 23 | Pgk1 | 1605 | 0.340 | 0.2057 | Yes |
| 24 | Pfkfb1 | 1818 | 0.313 | 0.1968 | Yes |
| 25 | Cd44 | 1920 | 0.301 | 0.1971 | Yes |
| 26 | Med24 | 1945 | 0.299 | 0.2041 | Yes |
| 27 | Spag4 | 2118 | 0.282 | 0.1977 | Yes |
| 28 | Ier3 | 2227 | 0.273 | 0.1966 | Yes |
| 29 | Sdhc | 2310 | 0.265 | 0.1975 | Yes |
| 30 | Cyb5a | 2377 | 0.259 | 0.1997 | Yes |
| 31 | Ecd | 2424 | 0.254 | 0.2034 | Yes |
| 32 | Egln3 | 2623 | 0.238 | 0.1934 | Yes |
| 33 | Slc25a13 | 2654 | 0.235 | 0.1979 | Yes |
| 34 | Mdh2 | 2669 | 0.234 | 0.2038 | Yes |
| 35 | B3gat3 | 2695 | 0.232 | 0.2087 | Yes |
| 36 | Ak4 | 2784 | 0.227 | 0.2079 | Yes |
| 37 | B4galt4 | 2801 | 0.226 | 0.2134 | Yes |
| 38 | Galk1 | 2883 | 0.220 | 0.2130 | No |
| 39 | Got2 | 2965 | 0.213 | 0.2124 | No |
| 40 | Mdh1 | 3128 | 0.199 | 0.2043 | No |
| 41 | Capn5 | 3311 | 0.187 | 0.1941 | No |
| 42 | Ext2 | 3546 | 0.167 | 0.1788 | No |
| 43 | Gclc | 3626 | 0.162 | 0.1768 | No |
| 44 | Xylt2 | 3720 | 0.154 | 0.1734 | No |
| 45 | Met | 3770 | 0.150 | 0.1737 | No |
| 46 | Gne | 3901 | 0.141 | 0.1667 | No |
| 47 | Glce | 4037 | 0.130 | 0.1589 | No |
| 48 | Prps1 | 4055 | 0.129 | 0.1613 | No |
| 49 | Gys1 | 4056 | 0.129 | 0.1652 | No |
| 50 | Lhpp | 4103 | 0.125 | 0.1650 | No |
| 51 | Zfp292 | 4130 | 0.123 | 0.1665 | No |
| 52 | P4ha1 | 4242 | 0.114 | 0.1603 | No |
| 53 | Hk2 | 4265 | 0.111 | 0.1618 | No |
| 54 | G6pdx | 4279 | 0.111 | 0.1640 | No |
| 55 | Hdlbp | 4301 | 0.109 | 0.1655 | No |
| 56 | Mpi | 4303 | 0.109 | 0.1687 | No |
| 57 | Alg1 | 4386 | 0.103 | 0.1647 | No |
| 58 | Aldoa | 4396 | 0.102 | 0.1670 | No |
| 59 | Pygb | 4500 | 0.095 | 0.1609 | No |
| 60 | Gnpda1 | 4553 | 0.091 | 0.1592 | No |
| 61 | Abcb6 | 4689 | 0.081 | 0.1499 | No |
| 62 | Pam | 4693 | 0.081 | 0.1521 | No |
| 63 | Hs2st1 | 4743 | 0.077 | 0.1501 | No |
| 64 | Ext1 | 4794 | 0.073 | 0.1480 | No |
| 65 | Mif | 4902 | 0.065 | 0.1407 | No |
| 66 | Irs2 | 4910 | 0.065 | 0.1420 | No |
| 67 | Ugp2 | 5001 | 0.058 | 0.1360 | No |
| 68 | Nsdhl | 5210 | 0.045 | 0.1192 | No |
| 69 | Gpc4 | 5269 | 0.040 | 0.1154 | No |
| 70 | Idua | 5364 | 0.033 | 0.1082 | No |
| 71 | Bpnt1 | 5473 | 0.024 | 0.0995 | No |
| 72 | Stmn1 | 5560 | 0.019 | 0.0925 | No |
| 73 | Cenpa | 5773 | 0.005 | 0.0742 | No |
| 74 | Arpp19 | 5775 | 0.004 | 0.0742 | No |
| 75 | Sdc2 | 5828 | 0.001 | 0.0697 | No |
| 76 | Polr3k | 6072 | -0.008 | 0.0488 | No |
| 77 | Fkbp4 | 6078 | -0.009 | 0.0486 | No |
| 78 | B3galt6 | 6200 | -0.017 | 0.0386 | No |
| 79 | Gpc1 | 6222 | -0.019 | 0.0373 | No |
| 80 | Plod2 | 6230 | -0.019 | 0.0373 | No |
| 81 | Taldo1 | 6231 | -0.019 | 0.0379 | No |
| 82 | Pfkp | 6310 | -0.026 | 0.0319 | No |
| 83 | Hax1 | 6347 | -0.029 | 0.0296 | No |
| 84 | Pgls | 6365 | -0.030 | 0.0290 | No |
| 85 | Psmc4 | 6468 | -0.036 | 0.0213 | No |
| 86 | Gmppb | 6507 | -0.040 | 0.0191 | No |
| 87 | Pgam1 | 6673 | -0.051 | 0.0063 | No |
| 88 | Hmmr | 6686 | -0.052 | 0.0068 | No |
| 89 | Kif20a | 6751 | -0.058 | 0.0030 | No |
| 90 | Fam162a | 7047 | -0.079 | -0.0203 | No |
| 91 | Pkm | 7086 | -0.082 | -0.0211 | No |
| 92 | Pygl | 7126 | -0.087 | -0.0218 | No |
| 93 | Cog2 | 7279 | -0.098 | -0.0321 | No |
| 94 | Plod1 | 7292 | -0.099 | -0.0301 | No |
| 95 | Ero1a | 7345 | -0.103 | -0.0316 | No |
| 96 | Rpe | 7512 | -0.116 | -0.0425 | No |
| 97 | Cdk1 | 7541 | -0.118 | -0.0414 | No |
| 98 | Ndufv3 | 7558 | -0.120 | -0.0391 | No |
| 99 | Tpi1 | 7676 | -0.127 | -0.0454 | No |
| 100 | Depdc1a | 8007 | -0.153 | -0.0695 | No |
| 101 | Srd5a3 | 8018 | -0.154 | -0.0657 | No |
| 102 | Txn1 | 8029 | -0.154 | -0.0619 | No |
| 103 | Galk2 | 8077 | -0.158 | -0.0612 | No |
| 104 | Copb2 | 8084 | -0.159 | -0.0569 | No |
| 105 | Dld | 8135 | -0.163 | -0.0563 | No |
| 106 | Gfus | 8335 | -0.179 | -0.0682 | No |
| 107 | Chpf2 | 8601 | -0.203 | -0.0851 | No |
| 108 | Gfpt1 | 8618 | -0.205 | -0.0803 | No |
| 109 | Akr1a1 | 8649 | -0.209 | -0.0765 | No |
| 110 | Hs6st2 | 8682 | -0.211 | -0.0729 | No |
| 111 | Tgfbi | 8712 | -0.213 | -0.0690 | No |
| 112 | B4galt1 | 8742 | -0.216 | -0.0649 | No |
| 113 | Slc37a4 | 8793 | -0.221 | -0.0626 | No |
| 114 | Angptl4 | 8842 | -0.225 | -0.0599 | No |
| 115 | Glrx | 9277 | -0.254 | -0.0900 | No |
| 116 | Ppp2cb | 9332 | -0.260 | -0.0868 | No |
| 117 | Nasp | 9334 | -0.260 | -0.0790 | No |
| 118 | Isg20 | 9510 | -0.279 | -0.0858 | No |
| 119 | Cited2 | 9569 | -0.285 | -0.0822 | No |
| 120 | Aldh9a1 | 9579 | -0.287 | -0.0742 | No |
| 121 | Pmm2 | 9620 | -0.292 | -0.0688 | No |
| 122 | Hspa5 | 9682 | -0.299 | -0.0651 | No |
| 123 | Agl | 9752 | -0.306 | -0.0618 | No |
| 124 | Aurka | 9767 | -0.308 | -0.0536 | No |
| 125 | Fut8 | 9894 | -0.320 | -0.0549 | No |
| 126 | Phka2 | 10036 | -0.336 | -0.0569 | No |
| 127 | Agrn | 10058 | -0.339 | -0.0485 | No |
| 128 | Qsox1 | 10064 | -0.340 | -0.0386 | No |
| 129 | Pdk3 | 10083 | -0.343 | -0.0297 | No |
| 130 | Cxcr4 | 10085 | -0.343 | -0.0194 | No |
| 131 | Rbck1 | 10103 | -0.344 | -0.0104 | No |
| 132 | Pcx | 10180 | -0.356 | -0.0062 | No |
| 133 | Gmppa | 10220 | -0.361 | 0.0014 | No |
| 134 | Gpc3 | 10335 | -0.378 | 0.0030 | No |
| 135 | Cln6 | 10741 | -0.441 | -0.0189 | No |
| 136 | Sod1 | 10801 | -0.451 | -0.0103 | No |
| 137 | Ldha | 11081 | -0.510 | -0.0191 | No |
| 138 | Kif2a | 11156 | -0.536 | -0.0093 | No |
| 139 | Sap30 | 11170 | -0.539 | 0.0060 | No |
| 140 | Pgm2 | 11600 | -1.059 | 0.0008 | No |