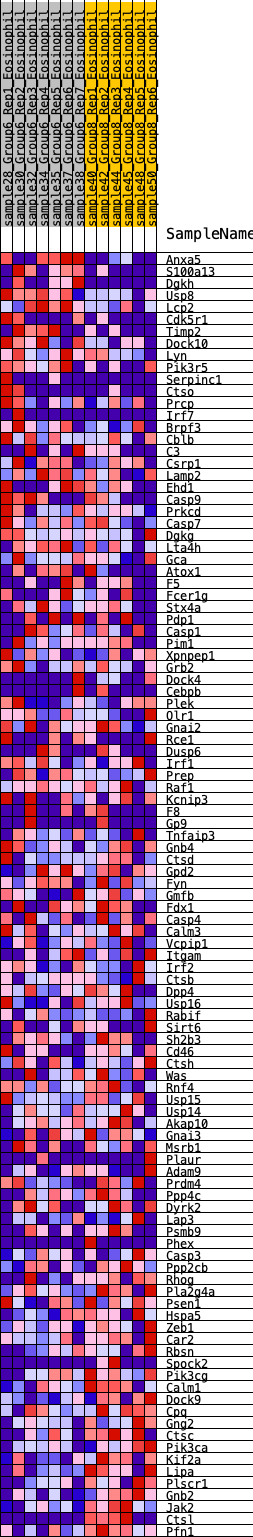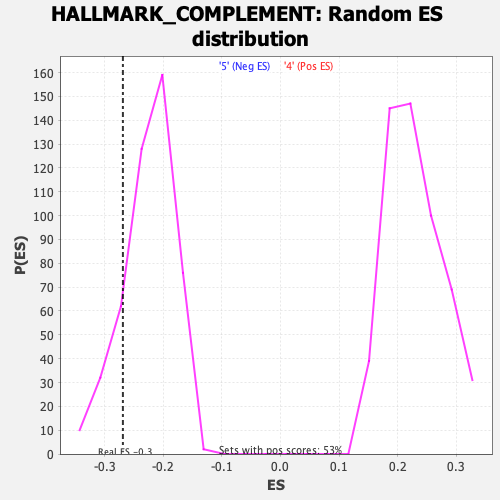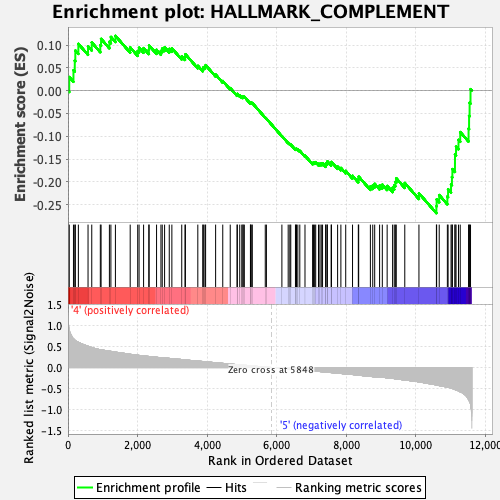
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group6_versus_Group8.Eosinophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Eosinophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | -0.26877958 |
| Normalized Enrichment Score (NES) | -1.19732 |
| Nominal p-value | 0.17910448 |
| FDR q-value | 0.47836274 |
| FWER p-Value | 0.819 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Anxa5 | 36 | 0.897 | 0.0299 | No |
| 2 | S100a13 | 157 | 0.690 | 0.0448 | No |
| 3 | Dgkh | 195 | 0.656 | 0.0657 | No |
| 4 | Usp8 | 211 | 0.641 | 0.0880 | No |
| 5 | Lcp2 | 298 | 0.601 | 0.1026 | No |
| 6 | Cdk5r1 | 576 | 0.508 | 0.0972 | No |
| 7 | Timp2 | 683 | 0.479 | 0.1056 | No |
| 8 | Dock10 | 929 | 0.425 | 0.0999 | No |
| 9 | Lyn | 951 | 0.421 | 0.1136 | No |
| 10 | Pik3r5 | 1190 | 0.391 | 0.1073 | No |
| 11 | Serpinc1 | 1233 | 0.386 | 0.1178 | No |
| 12 | Ctso | 1363 | 0.369 | 0.1202 | No |
| 13 | Prcp | 1788 | 0.316 | 0.0949 | No |
| 14 | Irf7 | 2004 | 0.292 | 0.0870 | No |
| 15 | Brpf3 | 2043 | 0.289 | 0.0943 | No |
| 16 | Cblb | 2173 | 0.278 | 0.0933 | No |
| 17 | C3 | 2325 | 0.264 | 0.0899 | No |
| 18 | Csrp1 | 2330 | 0.263 | 0.0992 | No |
| 19 | Lamp2 | 2546 | 0.245 | 0.0896 | No |
| 20 | Ehd1 | 2672 | 0.234 | 0.0873 | No |
| 21 | Casp9 | 2709 | 0.231 | 0.0926 | No |
| 22 | Prkcd | 2777 | 0.228 | 0.0952 | No |
| 23 | Casp7 | 2911 | 0.218 | 0.0916 | No |
| 24 | Dgkg | 2986 | 0.211 | 0.0930 | No |
| 25 | Lta4h | 3270 | 0.190 | 0.0754 | No |
| 26 | Gca | 3364 | 0.183 | 0.0740 | No |
| 27 | Atox1 | 3375 | 0.181 | 0.0798 | No |
| 28 | F5 | 3732 | 0.153 | 0.0545 | No |
| 29 | Fcer1g | 3874 | 0.143 | 0.0475 | No |
| 30 | Stx4a | 3891 | 0.142 | 0.0513 | No |
| 31 | Pdp1 | 3936 | 0.139 | 0.0526 | No |
| 32 | Casp1 | 3956 | 0.137 | 0.0560 | No |
| 33 | Pim1 | 4244 | 0.114 | 0.0352 | No |
| 34 | Xpnpep1 | 4449 | 0.098 | 0.0211 | No |
| 35 | Grb2 | 4664 | 0.083 | 0.0056 | No |
| 36 | Dock4 | 4860 | 0.068 | -0.0089 | No |
| 37 | Cebpb | 4868 | 0.067 | -0.0070 | No |
| 38 | Plek | 4937 | 0.063 | -0.0106 | No |
| 39 | Olr1 | 4996 | 0.059 | -0.0135 | No |
| 40 | Gnai2 | 5029 | 0.056 | -0.0142 | No |
| 41 | Rce1 | 5036 | 0.056 | -0.0127 | No |
| 42 | Dusp6 | 5069 | 0.053 | -0.0135 | No |
| 43 | Irf1 | 5245 | 0.043 | -0.0272 | No |
| 44 | Prep | 5246 | 0.042 | -0.0256 | No |
| 45 | Raf1 | 5282 | 0.039 | -0.0272 | No |
| 46 | Kcnip3 | 5297 | 0.038 | -0.0271 | No |
| 47 | F8 | 5671 | 0.011 | -0.0591 | No |
| 48 | Gp9 | 5704 | 0.008 | -0.0616 | No |
| 49 | Tnfaip3 | 6149 | -0.013 | -0.0997 | No |
| 50 | Gnb4 | 6330 | -0.027 | -0.1143 | No |
| 51 | Ctsd | 6377 | -0.031 | -0.1172 | No |
| 52 | Gpd2 | 6404 | -0.033 | -0.1183 | No |
| 53 | Fyn | 6539 | -0.042 | -0.1284 | No |
| 54 | Gmfb | 6552 | -0.043 | -0.1278 | No |
| 55 | Fdx1 | 6569 | -0.045 | -0.1276 | No |
| 56 | Casp4 | 6607 | -0.047 | -0.1291 | No |
| 57 | Calm3 | 6664 | -0.050 | -0.1321 | No |
| 58 | Vcpip1 | 6813 | -0.063 | -0.1426 | No |
| 59 | Itgam | 7032 | -0.079 | -0.1587 | No |
| 60 | Irf2 | 7059 | -0.080 | -0.1580 | No |
| 61 | Ctsb | 7090 | -0.083 | -0.1575 | No |
| 62 | Dpp4 | 7121 | -0.086 | -0.1570 | No |
| 63 | Usp16 | 7202 | -0.093 | -0.1605 | No |
| 64 | Rabif | 7242 | -0.096 | -0.1604 | No |
| 65 | Sirt6 | 7281 | -0.098 | -0.1601 | No |
| 66 | Sh2b3 | 7318 | -0.101 | -0.1595 | No |
| 67 | Cd46 | 7408 | -0.107 | -0.1633 | No |
| 68 | Ctsh | 7412 | -0.107 | -0.1596 | No |
| 69 | Was | 7449 | -0.110 | -0.1587 | No |
| 70 | Rnf4 | 7456 | -0.111 | -0.1552 | No |
| 71 | Usp15 | 7565 | -0.120 | -0.1601 | No |
| 72 | Usp14 | 7575 | -0.121 | -0.1565 | No |
| 73 | Akap10 | 7751 | -0.134 | -0.1667 | No |
| 74 | Gnai3 | 7845 | -0.142 | -0.1696 | No |
| 75 | Msrb1 | 7984 | -0.151 | -0.1760 | No |
| 76 | Plaur | 8180 | -0.167 | -0.1869 | No |
| 77 | Adam9 | 8346 | -0.181 | -0.1945 | No |
| 78 | Prdm4 | 8357 | -0.182 | -0.1887 | No |
| 79 | Ppp4c | 8693 | -0.211 | -0.2101 | No |
| 80 | Dyrk2 | 8761 | -0.218 | -0.2079 | No |
| 81 | Lap3 | 8817 | -0.223 | -0.2045 | No |
| 82 | Psmb9 | 8958 | -0.232 | -0.2081 | No |
| 83 | Phex | 9036 | -0.237 | -0.2061 | No |
| 84 | Casp3 | 9175 | -0.245 | -0.2091 | No |
| 85 | Ppp2cb | 9332 | -0.260 | -0.2131 | No |
| 86 | Rhog | 9379 | -0.264 | -0.2074 | No |
| 87 | Pla2g4a | 9411 | -0.269 | -0.2002 | No |
| 88 | Psen1 | 9437 | -0.272 | -0.1923 | No |
| 89 | Hspa5 | 9682 | -0.299 | -0.2026 | No |
| 90 | Zeb1 | 10089 | -0.344 | -0.2252 | No |
| 91 | Car2 | 10591 | -0.416 | -0.2535 | Yes |
| 92 | Rbsn | 10598 | -0.417 | -0.2387 | Yes |
| 93 | Spock2 | 10672 | -0.432 | -0.2291 | Yes |
| 94 | Pik3cg | 10906 | -0.470 | -0.2321 | Yes |
| 95 | Calm1 | 10927 | -0.473 | -0.2164 | Yes |
| 96 | Dock9 | 11012 | -0.491 | -0.2056 | Yes |
| 97 | Cpq | 11038 | -0.497 | -0.1895 | Yes |
| 98 | Gng2 | 11051 | -0.501 | -0.1722 | Yes |
| 99 | Ctsc | 11125 | -0.526 | -0.1592 | Yes |
| 100 | Pik3ca | 11126 | -0.526 | -0.1398 | Yes |
| 101 | Kif2a | 11156 | -0.536 | -0.1226 | Yes |
| 102 | Lipa | 11226 | -0.560 | -0.1080 | Yes |
| 103 | Plscr1 | 11276 | -0.581 | -0.0909 | Yes |
| 104 | Gnb2 | 11516 | -0.772 | -0.0833 | Yes |
| 105 | Jak2 | 11531 | -0.792 | -0.0554 | Yes |
| 106 | Ctsl | 11546 | -0.817 | -0.0266 | Yes |
| 107 | Pfn1 | 11572 | -0.869 | 0.0032 | Yes |