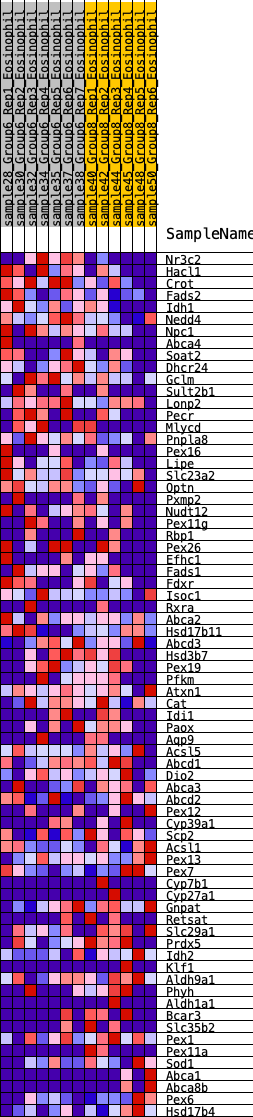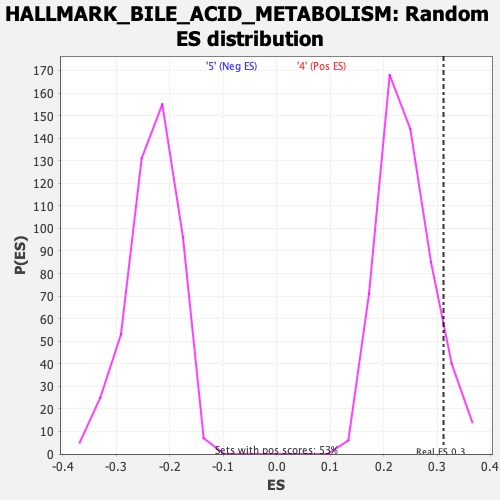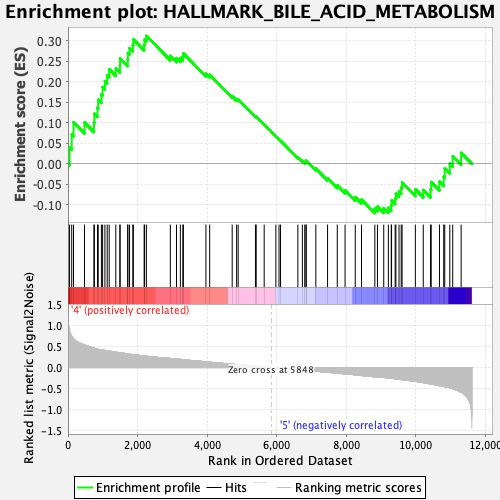
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group6_versus_Group8.Eosinophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Eosinophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.31219354 |
| Normalized Enrichment Score (NES) | 1.2929935 |
| Nominal p-value | 0.083333336 |
| FDR q-value | 0.47137642 |
| FWER p-Value | 0.704 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nr3c2 | 45 | 0.873 | 0.0396 | Yes |
| 2 | Hacl1 | 107 | 0.749 | 0.0716 | Yes |
| 3 | Crot | 159 | 0.687 | 0.1014 | Yes |
| 4 | Fads2 | 474 | 0.538 | 0.1010 | Yes |
| 5 | Idh1 | 745 | 0.462 | 0.1006 | Yes |
| 6 | Nedd4 | 758 | 0.458 | 0.1223 | Yes |
| 7 | Npc1 | 848 | 0.437 | 0.1364 | Yes |
| 8 | Abca4 | 868 | 0.435 | 0.1564 | Yes |
| 9 | Soat2 | 960 | 0.420 | 0.1694 | Yes |
| 10 | Dhcr24 | 995 | 0.415 | 0.1871 | Yes |
| 11 | Gclm | 1061 | 0.408 | 0.2018 | Yes |
| 12 | Sult2b1 | 1127 | 0.401 | 0.2161 | Yes |
| 13 | Lonp2 | 1184 | 0.393 | 0.2309 | Yes |
| 14 | Pecr | 1374 | 0.367 | 0.2328 | Yes |
| 15 | Mlycd | 1492 | 0.353 | 0.2402 | Yes |
| 16 | Pnpla8 | 1497 | 0.352 | 0.2574 | Yes |
| 17 | Pex16 | 1716 | 0.325 | 0.2547 | Yes |
| 18 | Lipe | 1721 | 0.325 | 0.2705 | Yes |
| 19 | Slc23a2 | 1769 | 0.318 | 0.2823 | Yes |
| 20 | Optn | 1864 | 0.308 | 0.2895 | Yes |
| 21 | Pxmp2 | 1878 | 0.307 | 0.3036 | Yes |
| 22 | Nudt12 | 2189 | 0.276 | 0.2905 | Yes |
| 23 | Pex11g | 2201 | 0.275 | 0.3032 | Yes |
| 24 | Rbp1 | 2253 | 0.269 | 0.3122 | Yes |
| 25 | Pex26 | 2942 | 0.215 | 0.2633 | No |
| 26 | Efhc1 | 3119 | 0.199 | 0.2579 | No |
| 27 | Fads1 | 3230 | 0.194 | 0.2581 | No |
| 28 | Fdxr | 3302 | 0.188 | 0.2612 | No |
| 29 | Isoc1 | 3317 | 0.186 | 0.2693 | No |
| 30 | Rxra | 3965 | 0.136 | 0.2200 | No |
| 31 | Abca2 | 4073 | 0.127 | 0.2171 | No |
| 32 | Hsd17b11 | 4720 | 0.079 | 0.1650 | No |
| 33 | Abcd3 | 4848 | 0.069 | 0.1574 | No |
| 34 | Hsd3b7 | 4892 | 0.066 | 0.1570 | No |
| 35 | Pex19 | 5389 | 0.031 | 0.1155 | No |
| 36 | Pfkm | 5410 | 0.028 | 0.1152 | No |
| 37 | Atxn1 | 5640 | 0.013 | 0.0960 | No |
| 38 | Cat | 5976 | -0.000 | 0.0670 | No |
| 39 | Idi1 | 6066 | -0.007 | 0.0596 | No |
| 40 | Paox | 6110 | -0.010 | 0.0564 | No |
| 41 | Aqp9 | 6608 | -0.047 | 0.0157 | No |
| 42 | Acsl5 | 6738 | -0.057 | 0.0073 | No |
| 43 | Abcd1 | 6806 | -0.063 | 0.0047 | No |
| 44 | Dio2 | 6843 | -0.067 | 0.0048 | No |
| 45 | Abca3 | 6847 | -0.067 | 0.0079 | No |
| 46 | Abcd2 | 7125 | -0.086 | -0.0118 | No |
| 47 | Pex12 | 7462 | -0.112 | -0.0353 | No |
| 48 | Cyp39a1 | 7741 | -0.133 | -0.0528 | No |
| 49 | Scp2 | 7965 | -0.150 | -0.0646 | No |
| 50 | Acsl1 | 8258 | -0.173 | -0.0814 | No |
| 51 | Pex13 | 8438 | -0.190 | -0.0874 | No |
| 52 | Pex7 | 8821 | -0.223 | -0.1094 | No |
| 53 | Cyp7b1 | 8896 | -0.228 | -0.1044 | No |
| 54 | Cyp27a1 | 9077 | -0.239 | -0.1081 | No |
| 55 | Gnpat | 9210 | -0.247 | -0.1073 | No |
| 56 | Retsat | 9293 | -0.256 | -0.1016 | No |
| 57 | Slc29a1 | 9302 | -0.256 | -0.0896 | No |
| 58 | Prdx5 | 9405 | -0.269 | -0.0850 | No |
| 59 | Idh2 | 9426 | -0.271 | -0.0733 | No |
| 60 | Klf1 | 9516 | -0.279 | -0.0671 | No |
| 61 | Aldh9a1 | 9579 | -0.287 | -0.0582 | No |
| 62 | Phyh | 9604 | -0.290 | -0.0458 | No |
| 63 | Aldh1a1 | 9986 | -0.330 | -0.0624 | No |
| 64 | Bcar3 | 10213 | -0.361 | -0.0640 | No |
| 65 | Slc35b2 | 10423 | -0.391 | -0.0627 | No |
| 66 | Pex1 | 10441 | -0.394 | -0.0445 | No |
| 67 | Pex11a | 10679 | -0.433 | -0.0435 | No |
| 68 | Sod1 | 10801 | -0.451 | -0.0315 | No |
| 69 | Abca1 | 10831 | -0.457 | -0.0113 | No |
| 70 | Abca8b | 10976 | -0.482 | 0.0003 | No |
| 71 | Pex6 | 11060 | -0.504 | 0.0182 | No |
| 72 | Hsd17b4 | 11302 | -0.589 | 0.0266 | No |