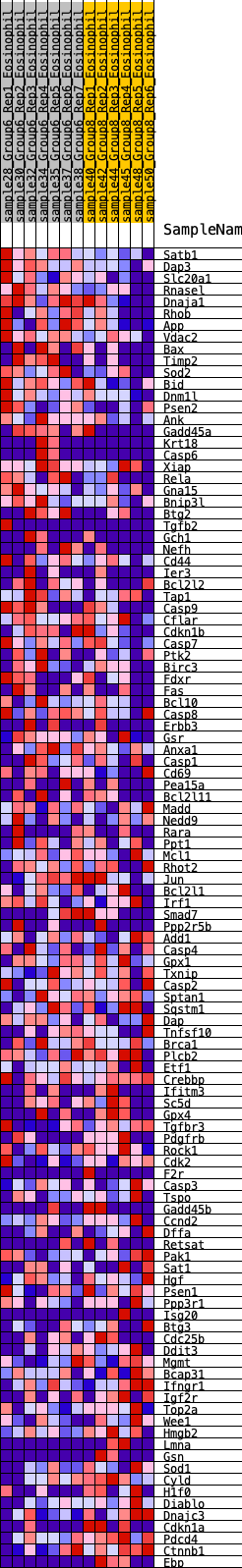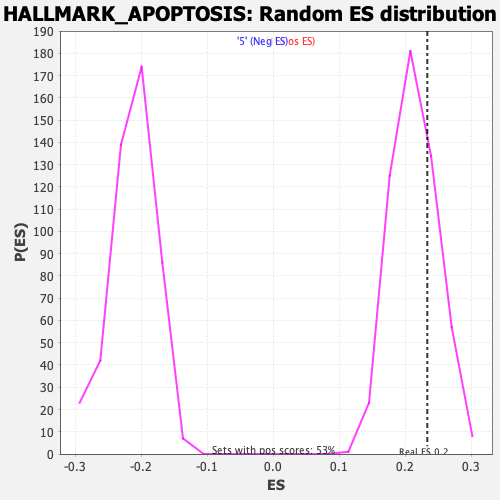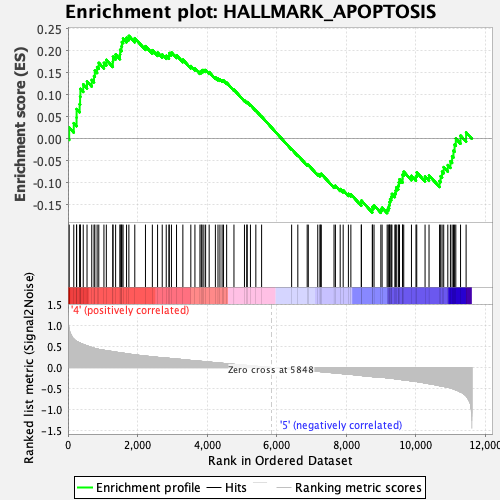
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group6_versus_Group8.Eosinophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Eosinophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | 0.23325372 |
| Normalized Enrichment Score (NES) | 1.0911984 |
| Nominal p-value | 0.28544423 |
| FDR q-value | 0.9352772 |
| FWER p-Value | 0.963 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Satb1 | 34 | 0.905 | 0.0254 | Yes |
| 2 | Dap3 | 166 | 0.679 | 0.0352 | Yes |
| 3 | Slc20a1 | 246 | 0.624 | 0.0479 | Yes |
| 4 | Rnasel | 248 | 0.623 | 0.0673 | Yes |
| 5 | Dnaja1 | 335 | 0.588 | 0.0783 | Yes |
| 6 | Rhob | 348 | 0.583 | 0.0955 | Yes |
| 7 | App | 356 | 0.581 | 0.1130 | Yes |
| 8 | Vdac2 | 438 | 0.551 | 0.1232 | Yes |
| 9 | Bax | 547 | 0.518 | 0.1301 | Yes |
| 10 | Timp2 | 683 | 0.479 | 0.1333 | Yes |
| 11 | Sod2 | 748 | 0.460 | 0.1421 | Yes |
| 12 | Bid | 772 | 0.455 | 0.1544 | Yes |
| 13 | Dnm1l | 837 | 0.439 | 0.1626 | Yes |
| 14 | Psen2 | 885 | 0.432 | 0.1720 | Yes |
| 15 | Ank | 1033 | 0.411 | 0.1721 | Yes |
| 16 | Gadd45a | 1102 | 0.403 | 0.1788 | Yes |
| 17 | Krt18 | 1289 | 0.378 | 0.1745 | Yes |
| 18 | Casp6 | 1293 | 0.378 | 0.1861 | Yes |
| 19 | Xiap | 1369 | 0.368 | 0.1911 | Yes |
| 20 | Rela | 1495 | 0.352 | 0.1912 | Yes |
| 21 | Gna15 | 1500 | 0.351 | 0.2019 | Yes |
| 22 | Bnip3l | 1539 | 0.346 | 0.2094 | Yes |
| 23 | Btg2 | 1550 | 0.345 | 0.2193 | Yes |
| 24 | Tgfb2 | 1583 | 0.341 | 0.2273 | Yes |
| 25 | Gch1 | 1683 | 0.329 | 0.2289 | Yes |
| 26 | Nefh | 1750 | 0.321 | 0.2333 | Yes |
| 27 | Cd44 | 1920 | 0.301 | 0.2280 | No |
| 28 | Ier3 | 2227 | 0.273 | 0.2099 | No |
| 29 | Bcl2l2 | 2425 | 0.254 | 0.2007 | No |
| 30 | Tap1 | 2575 | 0.242 | 0.1953 | No |
| 31 | Casp9 | 2709 | 0.231 | 0.1910 | No |
| 32 | Cflar | 2824 | 0.223 | 0.1881 | No |
| 33 | Cdkn1b | 2902 | 0.218 | 0.1882 | No |
| 34 | Casp7 | 2911 | 0.218 | 0.1943 | No |
| 35 | Ptk2 | 2974 | 0.212 | 0.1956 | No |
| 36 | Birc3 | 3120 | 0.199 | 0.1892 | No |
| 37 | Fdxr | 3302 | 0.188 | 0.1793 | No |
| 38 | Fas | 3531 | 0.168 | 0.1648 | No |
| 39 | Bcl10 | 3649 | 0.159 | 0.1596 | No |
| 40 | Casp8 | 3794 | 0.148 | 0.1517 | No |
| 41 | Erbb3 | 3832 | 0.146 | 0.1530 | No |
| 42 | Gsr | 3855 | 0.145 | 0.1557 | No |
| 43 | Anxa1 | 3906 | 0.141 | 0.1557 | No |
| 44 | Casp1 | 3956 | 0.137 | 0.1557 | No |
| 45 | Cd69 | 4058 | 0.129 | 0.1510 | No |
| 46 | Pea15a | 4237 | 0.114 | 0.1391 | No |
| 47 | Bcl2l11 | 4312 | 0.109 | 0.1361 | No |
| 48 | Madd | 4368 | 0.105 | 0.1346 | No |
| 49 | Nedd9 | 4430 | 0.100 | 0.1324 | No |
| 50 | Rara | 4469 | 0.097 | 0.1321 | No |
| 51 | Ppt1 | 4561 | 0.091 | 0.1270 | No |
| 52 | Mcl1 | 4771 | 0.074 | 0.1112 | No |
| 53 | Rhot2 | 5073 | 0.053 | 0.0867 | No |
| 54 | Jun | 5139 | 0.050 | 0.0826 | No |
| 55 | Bcl2l1 | 5152 | 0.049 | 0.0830 | No |
| 56 | Irf1 | 5245 | 0.043 | 0.0764 | No |
| 57 | Smad7 | 5400 | 0.029 | 0.0639 | No |
| 58 | Ppp2r5b | 5566 | 0.018 | 0.0501 | No |
| 59 | Add1 | 6429 | -0.034 | -0.0238 | No |
| 60 | Casp4 | 6607 | -0.047 | -0.0377 | No |
| 61 | Gpx1 | 6876 | -0.068 | -0.0589 | No |
| 62 | Txnip | 6908 | -0.071 | -0.0594 | No |
| 63 | Casp2 | 7180 | -0.091 | -0.0801 | No |
| 64 | Sptan1 | 7244 | -0.096 | -0.0826 | No |
| 65 | Sqstm1 | 7254 | -0.096 | -0.0803 | No |
| 66 | Dap | 7282 | -0.098 | -0.0796 | No |
| 67 | Tnfsf10 | 7648 | -0.125 | -0.1074 | No |
| 68 | Brca1 | 7688 | -0.128 | -0.1068 | No |
| 69 | Plcb2 | 7825 | -0.140 | -0.1142 | No |
| 70 | Etf1 | 7911 | -0.147 | -0.1170 | No |
| 71 | Crebbp | 8061 | -0.157 | -0.1250 | No |
| 72 | Ifitm3 | 8132 | -0.163 | -0.1260 | No |
| 73 | Sc5d | 8432 | -0.190 | -0.1461 | No |
| 74 | Gpx4 | 8435 | -0.190 | -0.1403 | No |
| 75 | Tgfbr3 | 8748 | -0.217 | -0.1607 | No |
| 76 | Pdgfrb | 8756 | -0.218 | -0.1545 | No |
| 77 | Rock1 | 8798 | -0.221 | -0.1511 | No |
| 78 | Cdk2 | 8994 | -0.235 | -0.1607 | No |
| 79 | F2r | 9030 | -0.237 | -0.1563 | No |
| 80 | Casp3 | 9175 | -0.245 | -0.1612 | No |
| 81 | Tspo | 9206 | -0.247 | -0.1561 | No |
| 82 | Gadd45b | 9232 | -0.249 | -0.1505 | No |
| 83 | Ccnd2 | 9245 | -0.251 | -0.1436 | No |
| 84 | Dffa | 9260 | -0.252 | -0.1370 | No |
| 85 | Retsat | 9293 | -0.256 | -0.1317 | No |
| 86 | Pak1 | 9312 | -0.258 | -0.1252 | No |
| 87 | Sat1 | 9397 | -0.268 | -0.1242 | No |
| 88 | Hgf | 9420 | -0.270 | -0.1176 | No |
| 89 | Psen1 | 9437 | -0.272 | -0.1105 | No |
| 90 | Ppp3r1 | 9494 | -0.278 | -0.1066 | No |
| 91 | Isg20 | 9510 | -0.279 | -0.0992 | No |
| 92 | Btg3 | 9530 | -0.280 | -0.0921 | No |
| 93 | Cdc25b | 9618 | -0.292 | -0.0905 | No |
| 94 | Ddit3 | 9619 | -0.292 | -0.0814 | No |
| 95 | Mgmt | 9652 | -0.295 | -0.0750 | No |
| 96 | Bcap31 | 9874 | -0.318 | -0.0842 | No |
| 97 | Ifngr1 | 10004 | -0.332 | -0.0850 | No |
| 98 | Igf2r | 10026 | -0.334 | -0.0764 | No |
| 99 | Top2a | 10265 | -0.368 | -0.0856 | No |
| 100 | Wee1 | 10379 | -0.386 | -0.0833 | No |
| 101 | Hmgb2 | 10682 | -0.433 | -0.0960 | No |
| 102 | Lmna | 10714 | -0.438 | -0.0850 | No |
| 103 | Gsn | 10754 | -0.443 | -0.0746 | No |
| 104 | Sod1 | 10801 | -0.451 | -0.0644 | No |
| 105 | Cyld | 10919 | -0.472 | -0.0598 | No |
| 106 | H1f0 | 10994 | -0.485 | -0.0511 | No |
| 107 | Diablo | 11042 | -0.499 | -0.0396 | No |
| 108 | Dnajc3 | 11082 | -0.511 | -0.0270 | No |
| 109 | Cdkn1a | 11114 | -0.522 | -0.0133 | No |
| 110 | Pdcd4 | 11149 | -0.534 | 0.0004 | No |
| 111 | Ctnnb1 | 11285 | -0.583 | 0.0070 | No |
| 112 | Ebp | 11445 | -0.675 | 0.0143 | No |