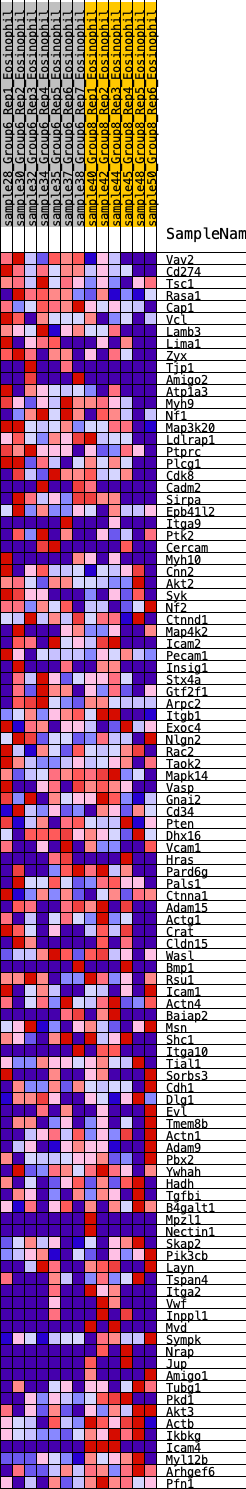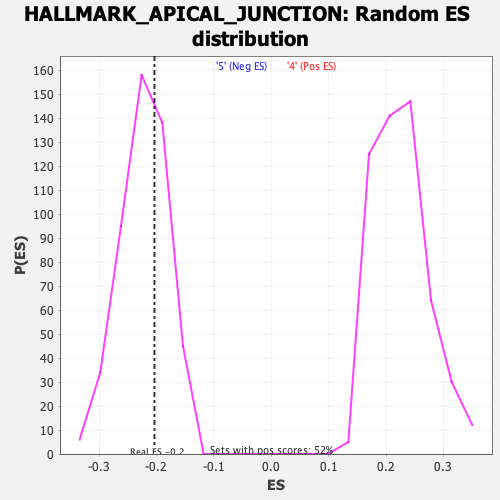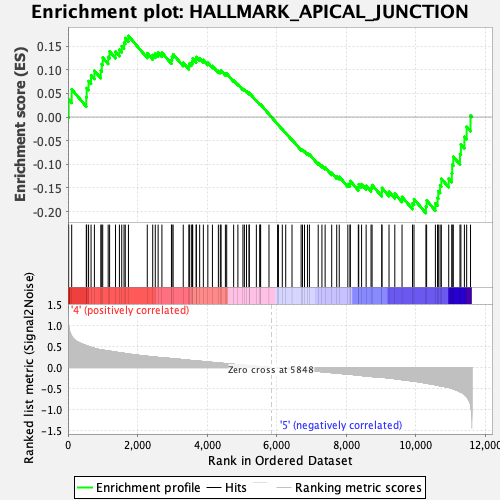
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group6_versus_Group8.Eosinophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Eosinophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_APICAL_JUNCTION |
| Enrichment Score (ES) | -0.20340465 |
| Normalized Enrichment Score (NES) | -0.91270787 |
| Nominal p-value | 0.6533613 |
| FDR q-value | 0.839804 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Vav2 | 19 | 1.010 | 0.0372 | No |
| 2 | Cd274 | 106 | 0.750 | 0.0586 | No |
| 3 | Tsc1 | 526 | 0.523 | 0.0423 | No |
| 4 | Rasa1 | 537 | 0.520 | 0.0614 | No |
| 5 | Cap1 | 591 | 0.503 | 0.0762 | No |
| 6 | Vcl | 663 | 0.483 | 0.0886 | No |
| 7 | Lamb3 | 759 | 0.458 | 0.0979 | No |
| 8 | Lima1 | 943 | 0.422 | 0.0983 | No |
| 9 | Zyx | 970 | 0.418 | 0.1121 | No |
| 10 | Tjp1 | 998 | 0.415 | 0.1257 | No |
| 11 | Amigo2 | 1155 | 0.397 | 0.1274 | No |
| 12 | Atp1a3 | 1196 | 0.390 | 0.1389 | No |
| 13 | Myh9 | 1366 | 0.368 | 0.1384 | No |
| 14 | Nf1 | 1478 | 0.355 | 0.1424 | No |
| 15 | Map3k20 | 1545 | 0.346 | 0.1500 | No |
| 16 | Ldlrap1 | 1611 | 0.338 | 0.1573 | No |
| 17 | Ptprc | 1646 | 0.334 | 0.1672 | No |
| 18 | Plcg1 | 1738 | 0.323 | 0.1717 | No |
| 19 | Cdk8 | 2279 | 0.267 | 0.1351 | No |
| 20 | Cadm2 | 2435 | 0.254 | 0.1313 | No |
| 21 | Sirpa | 2509 | 0.249 | 0.1346 | No |
| 22 | Epb41l2 | 2591 | 0.241 | 0.1368 | No |
| 23 | Itga9 | 2700 | 0.232 | 0.1363 | No |
| 24 | Ptk2 | 2974 | 0.212 | 0.1207 | No |
| 25 | Cercam | 2989 | 0.211 | 0.1276 | No |
| 26 | Myh10 | 3022 | 0.208 | 0.1329 | No |
| 27 | Cnn2 | 3313 | 0.187 | 0.1148 | No |
| 28 | Akt2 | 3476 | 0.173 | 0.1074 | No |
| 29 | Syk | 3488 | 0.171 | 0.1130 | No |
| 30 | Nf2 | 3545 | 0.167 | 0.1146 | No |
| 31 | Ctnnd1 | 3576 | 0.165 | 0.1183 | No |
| 32 | Map4k2 | 3585 | 0.164 | 0.1239 | No |
| 33 | Icam2 | 3685 | 0.157 | 0.1214 | No |
| 34 | Pecam1 | 3689 | 0.157 | 0.1271 | No |
| 35 | Insig1 | 3785 | 0.149 | 0.1246 | No |
| 36 | Stx4a | 3891 | 0.142 | 0.1209 | No |
| 37 | Gtf2f1 | 4020 | 0.132 | 0.1149 | No |
| 38 | Arpc2 | 4152 | 0.121 | 0.1082 | No |
| 39 | Itgb1 | 4323 | 0.108 | 0.0975 | No |
| 40 | Exoc4 | 4379 | 0.104 | 0.0967 | No |
| 41 | Nlgn2 | 4401 | 0.102 | 0.0988 | No |
| 42 | Rac2 | 4525 | 0.093 | 0.0917 | No |
| 43 | Taok2 | 4562 | 0.091 | 0.0921 | No |
| 44 | Mapk14 | 4762 | 0.075 | 0.0777 | No |
| 45 | Vasp | 4884 | 0.067 | 0.0697 | No |
| 46 | Gnai2 | 5029 | 0.056 | 0.0594 | No |
| 47 | Cd34 | 5071 | 0.053 | 0.0578 | No |
| 48 | Pten | 5132 | 0.050 | 0.0545 | No |
| 49 | Dhx16 | 5207 | 0.045 | 0.0498 | No |
| 50 | Vcam1 | 5209 | 0.045 | 0.0515 | No |
| 51 | Hras | 5415 | 0.028 | 0.0347 | No |
| 52 | Pard6g | 5514 | 0.022 | 0.0271 | No |
| 53 | Pals1 | 5523 | 0.021 | 0.0272 | No |
| 54 | Ctnna1 | 5538 | 0.020 | 0.0267 | No |
| 55 | Adam15 | 5778 | 0.004 | 0.0061 | No |
| 56 | Actg1 | 6025 | -0.004 | -0.0151 | No |
| 57 | Crat | 6050 | -0.006 | -0.0170 | No |
| 58 | Cldn15 | 6161 | -0.014 | -0.0260 | No |
| 59 | Wasl | 6259 | -0.022 | -0.0336 | No |
| 60 | Bmp1 | 6440 | -0.035 | -0.0479 | No |
| 61 | Rsu1 | 6702 | -0.054 | -0.0685 | No |
| 62 | Icam1 | 6741 | -0.058 | -0.0696 | No |
| 63 | Actn4 | 6799 | -0.062 | -0.0722 | No |
| 64 | Baiap2 | 6884 | -0.069 | -0.0768 | No |
| 65 | Msn | 6940 | -0.072 | -0.0788 | No |
| 66 | Shc1 | 7193 | -0.092 | -0.0972 | No |
| 67 | Itga10 | 7300 | -0.099 | -0.1026 | No |
| 68 | Tial1 | 7392 | -0.106 | -0.1064 | No |
| 69 | Sorbs3 | 7581 | -0.121 | -0.1181 | No |
| 70 | Cdh1 | 7725 | -0.132 | -0.1254 | No |
| 71 | Dlg1 | 7798 | -0.138 | -0.1264 | No |
| 72 | Evl | 8045 | -0.156 | -0.1418 | No |
| 73 | Tmem8b | 8107 | -0.161 | -0.1409 | No |
| 74 | Actn1 | 8115 | -0.161 | -0.1353 | No |
| 75 | Adam9 | 8346 | -0.181 | -0.1483 | No |
| 76 | Pbx2 | 8358 | -0.182 | -0.1423 | No |
| 77 | Ywhah | 8440 | -0.190 | -0.1420 | No |
| 78 | Hadh | 8571 | -0.201 | -0.1456 | No |
| 79 | Tgfbi | 8712 | -0.213 | -0.1496 | No |
| 80 | B4galt1 | 8742 | -0.216 | -0.1438 | No |
| 81 | Mpzl1 | 9022 | -0.237 | -0.1589 | No |
| 82 | Nectin1 | 9025 | -0.237 | -0.1499 | No |
| 83 | Skap2 | 9229 | -0.249 | -0.1580 | No |
| 84 | Pik3cb | 9395 | -0.268 | -0.1621 | No |
| 85 | Layn | 9603 | -0.290 | -0.1689 | No |
| 86 | Tspan4 | 9904 | -0.321 | -0.1826 | No |
| 87 | Itga2 | 9947 | -0.327 | -0.1737 | No |
| 88 | Vwf | 10290 | -0.372 | -0.1891 | Yes |
| 89 | Inppl1 | 10310 | -0.374 | -0.1764 | Yes |
| 90 | Mvd | 10562 | -0.413 | -0.1823 | Yes |
| 91 | Sympk | 10624 | -0.423 | -0.1713 | Yes |
| 92 | Nrap | 10646 | -0.427 | -0.1567 | Yes |
| 93 | Jup | 10697 | -0.436 | -0.1443 | Yes |
| 94 | Amigo1 | 10733 | -0.440 | -0.1304 | Yes |
| 95 | Tubg1 | 10945 | -0.476 | -0.1304 | Yes |
| 96 | Pkd1 | 11034 | -0.496 | -0.1190 | Yes |
| 97 | Akt3 | 11045 | -0.500 | -0.1007 | Yes |
| 98 | Actb | 11077 | -0.509 | -0.0838 | Yes |
| 99 | Ikbkg | 11269 | -0.579 | -0.0781 | Yes |
| 100 | Icam4 | 11295 | -0.589 | -0.0577 | Yes |
| 101 | Myl12b | 11396 | -0.640 | -0.0418 | Yes |
| 102 | Arhgef6 | 11461 | -0.694 | -0.0206 | Yes |
| 103 | Pfn1 | 11572 | -0.869 | 0.0032 | Yes |