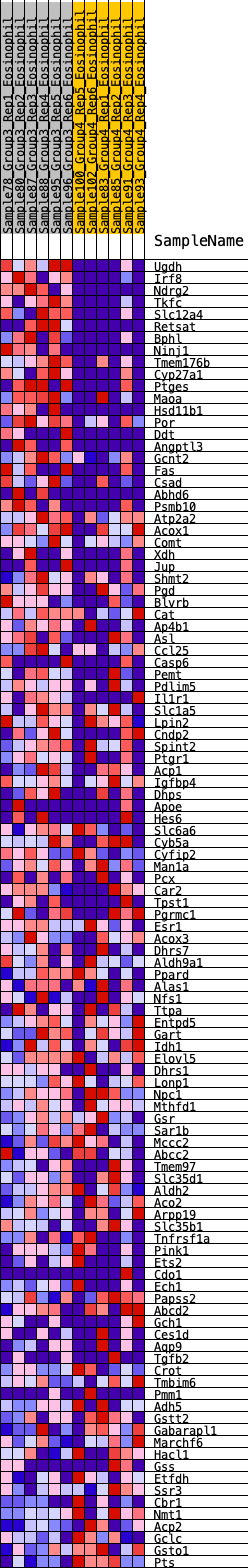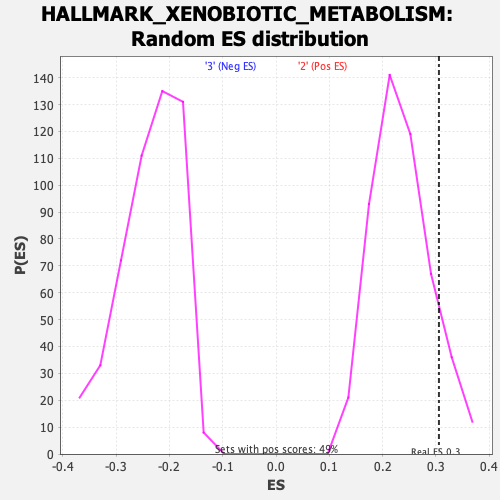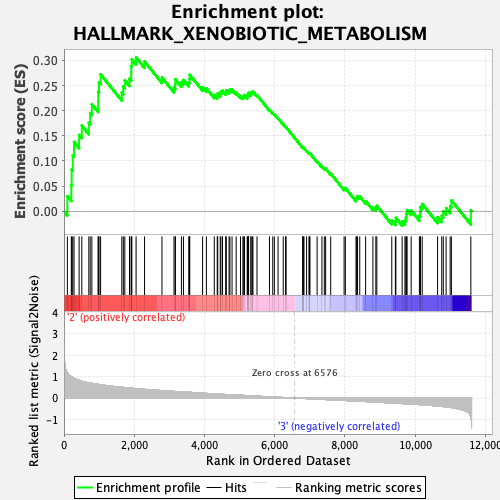
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group3_versus_Group4.Eosinophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | 0.30582175 |
| Normalized Enrichment Score (NES) | 1.2913337 |
| Nominal p-value | 0.12474438 |
| FDR q-value | 0.20958963 |
| FWER p-Value | 0.683 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ugdh | 96 | 1.194 | 0.0299 | Yes |
| 2 | Irf8 | 208 | 0.996 | 0.0521 | Yes |
| 3 | Ndrg2 | 219 | 0.986 | 0.0828 | Yes |
| 4 | Tkfc | 249 | 0.961 | 0.1111 | Yes |
| 5 | Slc12a4 | 285 | 0.929 | 0.1378 | Yes |
| 6 | Retsat | 431 | 0.827 | 0.1517 | Yes |
| 7 | Bphl | 507 | 0.783 | 0.1702 | Yes |
| 8 | Ninj1 | 707 | 0.714 | 0.1758 | Yes |
| 9 | Tmem176b | 746 | 0.700 | 0.1949 | Yes |
| 10 | Cyp27a1 | 794 | 0.686 | 0.2128 | Yes |
| 11 | Ptges | 974 | 0.637 | 0.2176 | Yes |
| 12 | Maoa | 976 | 0.637 | 0.2379 | Yes |
| 13 | Hsd11b1 | 999 | 0.631 | 0.2562 | Yes |
| 14 | Por | 1042 | 0.618 | 0.2724 | Yes |
| 15 | Ddt | 1647 | 0.503 | 0.2360 | Yes |
| 16 | Angptl3 | 1690 | 0.495 | 0.2482 | Yes |
| 17 | Gcnt2 | 1732 | 0.488 | 0.2603 | Yes |
| 18 | Fas | 1865 | 0.467 | 0.2637 | Yes |
| 19 | Csad | 1915 | 0.460 | 0.2742 | Yes |
| 20 | Abhd6 | 1916 | 0.460 | 0.2889 | Yes |
| 21 | Psmb10 | 1932 | 0.455 | 0.3022 | Yes |
| 22 | Atp2a2 | 2053 | 0.438 | 0.3058 | Yes |
| 23 | Acox1 | 2294 | 0.405 | 0.2979 | No |
| 24 | Comt | 2790 | 0.347 | 0.2660 | No |
| 25 | Xdh | 3133 | 0.309 | 0.2461 | No |
| 26 | Jup | 3169 | 0.305 | 0.2529 | No |
| 27 | Shmt2 | 3172 | 0.304 | 0.2624 | No |
| 28 | Pgd | 3344 | 0.290 | 0.2568 | No |
| 29 | Blvrb | 3402 | 0.284 | 0.2610 | No |
| 30 | Cat | 3553 | 0.267 | 0.2565 | No |
| 31 | Ap4b1 | 3575 | 0.266 | 0.2632 | No |
| 32 | Asl | 3581 | 0.265 | 0.2713 | No |
| 33 | Ccl25 | 3947 | 0.225 | 0.2468 | No |
| 34 | Casp6 | 4055 | 0.216 | 0.2444 | No |
| 35 | Pemt | 4280 | 0.191 | 0.2310 | No |
| 36 | Pdlim5 | 4364 | 0.183 | 0.2297 | No |
| 37 | Il1r1 | 4378 | 0.182 | 0.2344 | No |
| 38 | Slc1a5 | 4450 | 0.178 | 0.2339 | No |
| 39 | Lpin2 | 4469 | 0.176 | 0.2380 | No |
| 40 | Cndp2 | 4515 | 0.173 | 0.2396 | No |
| 41 | Spint2 | 4605 | 0.163 | 0.2371 | No |
| 42 | Ptgr1 | 4623 | 0.162 | 0.2408 | No |
| 43 | Acp1 | 4701 | 0.154 | 0.2390 | No |
| 44 | Igfbp4 | 4725 | 0.152 | 0.2419 | No |
| 45 | Dhps | 4786 | 0.148 | 0.2414 | No |
| 46 | Apoe | 4903 | 0.136 | 0.2357 | No |
| 47 | Hes6 | 5027 | 0.129 | 0.2291 | No |
| 48 | Slc6a6 | 5095 | 0.124 | 0.2273 | No |
| 49 | Cyb5a | 5119 | 0.121 | 0.2291 | No |
| 50 | Cyfip2 | 5141 | 0.117 | 0.2311 | No |
| 51 | Man1a | 5219 | 0.112 | 0.2280 | No |
| 52 | Pcx | 5224 | 0.111 | 0.2312 | No |
| 53 | Car2 | 5251 | 0.109 | 0.2324 | No |
| 54 | Tpst1 | 5257 | 0.108 | 0.2354 | No |
| 55 | Pgrmc1 | 5322 | 0.103 | 0.2332 | No |
| 56 | Esr1 | 5327 | 0.103 | 0.2361 | No |
| 57 | Acox3 | 5363 | 0.100 | 0.2362 | No |
| 58 | Dhrs7 | 5381 | 0.098 | 0.2379 | No |
| 59 | Aldh9a1 | 5497 | 0.086 | 0.2307 | No |
| 60 | Ppard | 5852 | 0.057 | 0.2017 | No |
| 61 | Alas1 | 5945 | 0.049 | 0.1953 | No |
| 62 | Nfs1 | 5991 | 0.043 | 0.1927 | No |
| 63 | Ttpa | 6096 | 0.036 | 0.1849 | No |
| 64 | Entpd5 | 6244 | 0.024 | 0.1728 | No |
| 65 | Gart | 6311 | 0.019 | 0.1677 | No |
| 66 | Idh1 | 6325 | 0.017 | 0.1671 | No |
| 67 | Elovl5 | 6800 | -0.016 | 0.1264 | No |
| 68 | Dhrs1 | 6801 | -0.016 | 0.1269 | No |
| 69 | Lonp1 | 6828 | -0.019 | 0.1253 | No |
| 70 | Npc1 | 6830 | -0.019 | 0.1258 | No |
| 71 | Mthfd1 | 6909 | -0.027 | 0.1199 | No |
| 72 | Gsr | 6977 | -0.033 | 0.1151 | No |
| 73 | Sar1b | 6991 | -0.034 | 0.1151 | No |
| 74 | Mccc2 | 7005 | -0.035 | 0.1151 | No |
| 75 | Abcc2 | 7209 | -0.052 | 0.0991 | No |
| 76 | Tmem97 | 7346 | -0.063 | 0.0893 | No |
| 77 | Slc35d1 | 7414 | -0.068 | 0.0857 | No |
| 78 | Aldh2 | 7451 | -0.071 | 0.0848 | No |
| 79 | Aco2 | 7596 | -0.081 | 0.0749 | No |
| 80 | Arpp19 | 7977 | -0.112 | 0.0455 | No |
| 81 | Slc35b1 | 8013 | -0.116 | 0.0461 | No |
| 82 | Tnfrsf1a | 8316 | -0.143 | 0.0244 | No |
| 83 | Pink1 | 8335 | -0.144 | 0.0275 | No |
| 84 | Ets2 | 8360 | -0.146 | 0.0300 | No |
| 85 | Cdo1 | 8423 | -0.151 | 0.0295 | No |
| 86 | Ech1 | 8590 | -0.165 | 0.0203 | No |
| 87 | Papss2 | 8798 | -0.182 | 0.0082 | No |
| 88 | Abcd2 | 8881 | -0.189 | 0.0071 | No |
| 89 | Gch1 | 8909 | -0.192 | 0.0109 | No |
| 90 | Ces1d | 9335 | -0.231 | -0.0186 | No |
| 91 | Aqp9 | 9438 | -0.242 | -0.0198 | No |
| 92 | Tgfb2 | 9452 | -0.243 | -0.0131 | No |
| 93 | Crot | 9631 | -0.260 | -0.0203 | No |
| 94 | Tmbim6 | 9710 | -0.267 | -0.0185 | No |
| 95 | Pmm1 | 9737 | -0.270 | -0.0121 | No |
| 96 | Adh5 | 9752 | -0.270 | -0.0047 | No |
| 97 | Gstt2 | 9771 | -0.272 | 0.0025 | No |
| 98 | Gabarapl1 | 9887 | -0.285 | 0.0016 | No |
| 99 | Marchf6 | 10122 | -0.307 | -0.0089 | No |
| 100 | Hacl1 | 10147 | -0.310 | -0.0011 | No |
| 101 | Gss | 10151 | -0.310 | 0.0086 | No |
| 102 | Etfdh | 10202 | -0.315 | 0.0143 | No |
| 103 | Ssr3 | 10640 | -0.376 | -0.0116 | No |
| 104 | Cbr1 | 10758 | -0.391 | -0.0093 | No |
| 105 | Nmt1 | 10802 | -0.399 | -0.0002 | No |
| 106 | Acp2 | 10882 | -0.414 | 0.0062 | No |
| 107 | Gclc | 10998 | -0.438 | 0.0102 | No |
| 108 | Gsto1 | 11033 | -0.446 | 0.0215 | No |
| 109 | Pts | 11587 | -0.889 | 0.0019 | No |