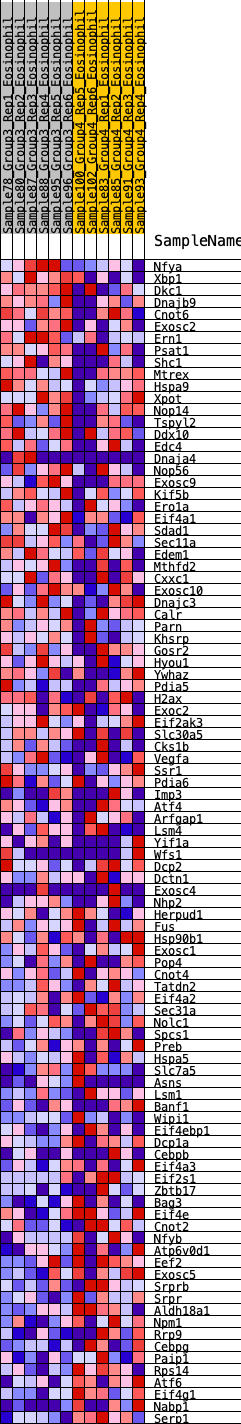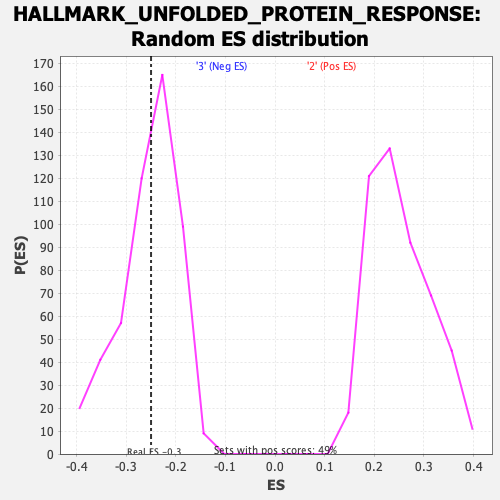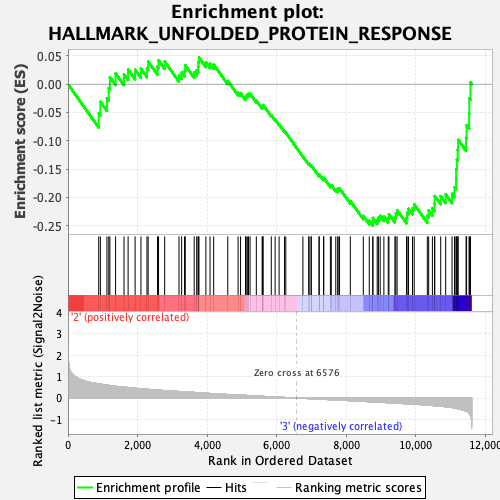
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group3_versus_Group4.Eosinophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | -0.25001004 |
| Normalized Enrichment Score (NES) | -0.98974603 |
| Nominal p-value | 0.45792565 |
| FDR q-value | 0.55173075 |
| FWER p-Value | 0.976 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nfya | 883 | 0.665 | -0.0516 | No |
| 2 | Xbp1 | 931 | 0.652 | -0.0310 | No |
| 3 | Dkc1 | 1119 | 0.600 | -0.0246 | No |
| 4 | Dnajb9 | 1172 | 0.590 | -0.0069 | No |
| 5 | Cnot6 | 1204 | 0.585 | 0.0125 | No |
| 6 | Exosc2 | 1368 | 0.548 | 0.0191 | No |
| 7 | Ern1 | 1610 | 0.509 | 0.0174 | No |
| 8 | Psat1 | 1728 | 0.489 | 0.0257 | No |
| 9 | Shc1 | 1929 | 0.457 | 0.0256 | No |
| 10 | Mtrex | 2097 | 0.432 | 0.0274 | No |
| 11 | Hspa9 | 2270 | 0.407 | 0.0278 | No |
| 12 | Xpot | 2304 | 0.403 | 0.0402 | No |
| 13 | Nop14 | 2573 | 0.370 | 0.0308 | No |
| 14 | Tspyl2 | 2603 | 0.366 | 0.0422 | No |
| 15 | Ddx10 | 2779 | 0.348 | 0.0401 | No |
| 16 | Edc4 | 3191 | 0.303 | 0.0158 | No |
| 17 | Dnaja4 | 3267 | 0.299 | 0.0206 | No |
| 18 | Nop56 | 3351 | 0.289 | 0.0243 | No |
| 19 | Exosc9 | 3371 | 0.287 | 0.0335 | No |
| 20 | Kif5b | 3628 | 0.260 | 0.0211 | No |
| 21 | Ero1a | 3699 | 0.253 | 0.0246 | No |
| 22 | Eif4a1 | 3742 | 0.249 | 0.0304 | No |
| 23 | Sdad1 | 3746 | 0.249 | 0.0395 | No |
| 24 | Sec11a | 3765 | 0.247 | 0.0473 | No |
| 25 | Edem1 | 3964 | 0.223 | 0.0385 | No |
| 26 | Mthfd2 | 4083 | 0.212 | 0.0362 | No |
| 27 | Cxxc1 | 4188 | 0.201 | 0.0348 | No |
| 28 | Exosc10 | 4593 | 0.164 | 0.0059 | No |
| 29 | Dnajc3 | 4890 | 0.137 | -0.0146 | No |
| 30 | Calr | 4961 | 0.134 | -0.0156 | No |
| 31 | Parn | 5103 | 0.123 | -0.0232 | No |
| 32 | Khsrp | 5121 | 0.121 | -0.0201 | No |
| 33 | Gosr2 | 5155 | 0.117 | -0.0186 | No |
| 34 | Hyou1 | 5189 | 0.114 | -0.0171 | No |
| 35 | Ywhaz | 5234 | 0.111 | -0.0168 | No |
| 36 | Pdia5 | 5414 | 0.095 | -0.0288 | No |
| 37 | H2ax | 5581 | 0.079 | -0.0402 | No |
| 38 | Exoc2 | 5593 | 0.078 | -0.0382 | No |
| 39 | Eif2ak3 | 5611 | 0.076 | -0.0368 | No |
| 40 | Slc30a5 | 5844 | 0.058 | -0.0548 | No |
| 41 | Cks1b | 5955 | 0.048 | -0.0626 | No |
| 42 | Vegfa | 6069 | 0.038 | -0.0709 | No |
| 43 | Ssr1 | 6225 | 0.025 | -0.0834 | No |
| 44 | Pdia6 | 6262 | 0.022 | -0.0857 | No |
| 45 | Imp3 | 6753 | -0.012 | -0.1278 | No |
| 46 | Atf4 | 6921 | -0.028 | -0.1413 | No |
| 47 | Arfgap1 | 6930 | -0.029 | -0.1408 | No |
| 48 | Lsm4 | 6989 | -0.034 | -0.1446 | No |
| 49 | Yif1a | 6993 | -0.035 | -0.1436 | No |
| 50 | Wfs1 | 7218 | -0.054 | -0.1610 | No |
| 51 | Dcp2 | 7221 | -0.054 | -0.1591 | No |
| 52 | Dctn1 | 7347 | -0.063 | -0.1676 | No |
| 53 | Exosc4 | 7353 | -0.063 | -0.1657 | No |
| 54 | Nhp2 | 7543 | -0.078 | -0.1791 | No |
| 55 | Herpud1 | 7569 | -0.080 | -0.1783 | No |
| 56 | Fus | 7698 | -0.090 | -0.1860 | No |
| 57 | Hsp90b1 | 7755 | -0.094 | -0.1873 | No |
| 58 | Exosc1 | 7764 | -0.095 | -0.1844 | No |
| 59 | Pop4 | 7806 | -0.098 | -0.1843 | No |
| 60 | Cnot4 | 8116 | -0.125 | -0.2064 | No |
| 61 | Tatdn2 | 8489 | -0.155 | -0.2328 | No |
| 62 | Eif4a2 | 8661 | -0.171 | -0.2412 | No |
| 63 | Sec31a | 8763 | -0.180 | -0.2432 | Yes |
| 64 | Nolc1 | 8764 | -0.180 | -0.2364 | Yes |
| 65 | Spcs1 | 8889 | -0.190 | -0.2400 | Yes |
| 66 | Preb | 8924 | -0.193 | -0.2357 | Yes |
| 67 | Hspa5 | 8975 | -0.197 | -0.2326 | Yes |
| 68 | Slc7a5 | 9081 | -0.208 | -0.2339 | Yes |
| 69 | Asns | 9206 | -0.220 | -0.2363 | Yes |
| 70 | Lsm1 | 9227 | -0.222 | -0.2297 | Yes |
| 71 | Banf1 | 9398 | -0.236 | -0.2355 | Yes |
| 72 | Wipi1 | 9424 | -0.240 | -0.2286 | Yes |
| 73 | Eif4ebp1 | 9461 | -0.244 | -0.2226 | Yes |
| 74 | Dcp1a | 9732 | -0.269 | -0.2358 | Yes |
| 75 | Cebpb | 9748 | -0.270 | -0.2269 | Yes |
| 76 | Eif4a3 | 9790 | -0.274 | -0.2202 | Yes |
| 77 | Eif2s1 | 9905 | -0.287 | -0.2192 | Yes |
| 78 | Zbtb17 | 9953 | -0.291 | -0.2123 | Yes |
| 79 | Bag3 | 10330 | -0.331 | -0.2325 | Yes |
| 80 | Eif4e | 10369 | -0.334 | -0.2232 | Yes |
| 81 | Cnot2 | 10476 | -0.351 | -0.2191 | Yes |
| 82 | Nfyb | 10538 | -0.360 | -0.2108 | Yes |
| 83 | Atp6v0d1 | 10544 | -0.361 | -0.1977 | Yes |
| 84 | Eef2 | 10715 | -0.386 | -0.1979 | Yes |
| 85 | Exosc5 | 10858 | -0.410 | -0.1947 | Yes |
| 86 | Srprb | 11041 | -0.448 | -0.1936 | Yes |
| 87 | Srpr | 11112 | -0.466 | -0.1821 | Yes |
| 88 | Aldh18a1 | 11157 | -0.477 | -0.1679 | Yes |
| 89 | Npm1 | 11160 | -0.478 | -0.1500 | Yes |
| 90 | Rrp9 | 11180 | -0.487 | -0.1333 | Yes |
| 91 | Cebpg | 11204 | -0.501 | -0.1164 | Yes |
| 92 | Paip1 | 11216 | -0.503 | -0.0983 | Yes |
| 93 | Rps14 | 11444 | -0.615 | -0.0948 | Yes |
| 94 | Atf6 | 11459 | -0.625 | -0.0724 | Yes |
| 95 | Eif4g1 | 11533 | -0.715 | -0.0518 | Yes |
| 96 | Nabp1 | 11538 | -0.723 | -0.0248 | Yes |
| 97 | Serp1 | 11572 | -0.818 | 0.0032 | Yes |