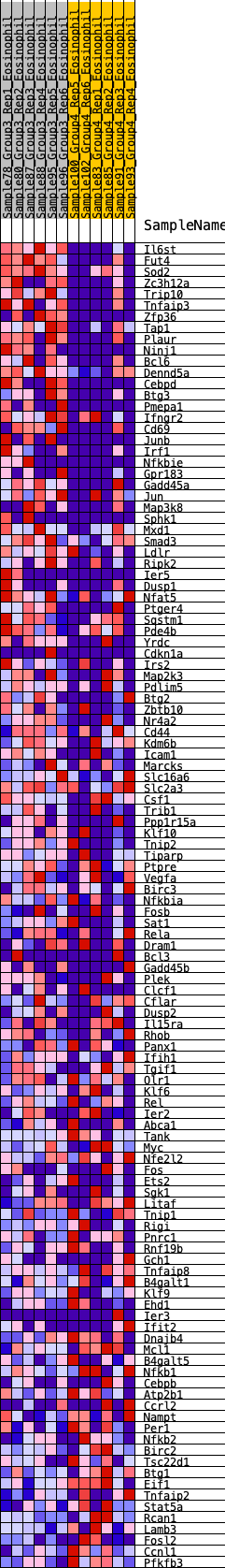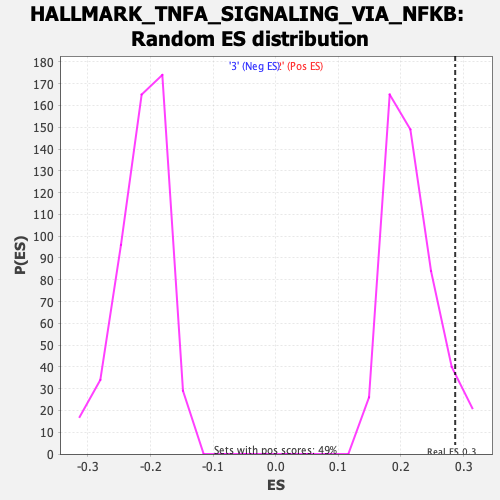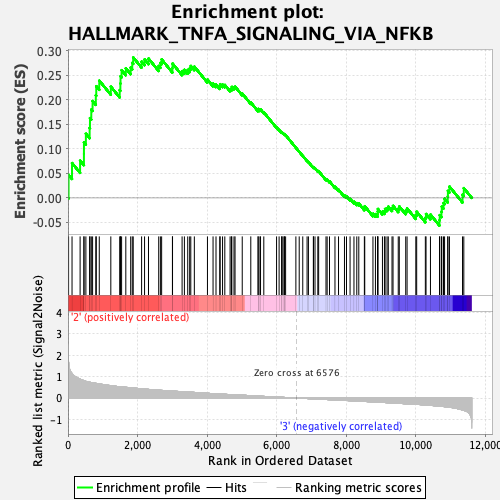
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group3_versus_Group4.Eosinophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_TNFA_SIGNALING_VIA_NFKB |
| Enrichment Score (ES) | 0.28666508 |
| Normalized Enrichment Score (NES) | 1.328969 |
| Nominal p-value | 0.068041235 |
| FDR q-value | 0.23043737 |
| FWER p-Value | 0.598 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Il6st | 17 | 1.696 | 0.0471 | Yes |
| 2 | Fut4 | 116 | 1.137 | 0.0711 | Yes |
| 3 | Sod2 | 348 | 0.880 | 0.0763 | Yes |
| 4 | Zc3h12a | 458 | 0.814 | 0.0901 | Yes |
| 5 | Trip10 | 459 | 0.813 | 0.1134 | Yes |
| 6 | Tnfaip3 | 514 | 0.779 | 0.1310 | Yes |
| 7 | Zfp36 | 622 | 0.735 | 0.1428 | Yes |
| 8 | Tap1 | 632 | 0.732 | 0.1630 | Yes |
| 9 | Plaur | 670 | 0.723 | 0.1804 | Yes |
| 10 | Ninj1 | 707 | 0.714 | 0.1978 | Yes |
| 11 | Bcl6 | 800 | 0.684 | 0.2094 | Yes |
| 12 | Dennd5a | 813 | 0.682 | 0.2279 | Yes |
| 13 | Cebpd | 899 | 0.660 | 0.2394 | Yes |
| 14 | Btg3 | 1231 | 0.577 | 0.2271 | Yes |
| 15 | Pmepa1 | 1487 | 0.529 | 0.2200 | Yes |
| 16 | Ifngr2 | 1506 | 0.525 | 0.2335 | Yes |
| 17 | Cd69 | 1510 | 0.525 | 0.2483 | Yes |
| 18 | Junb | 1541 | 0.521 | 0.2606 | Yes |
| 19 | Irf1 | 1662 | 0.501 | 0.2645 | Yes |
| 20 | Nfkbie | 1802 | 0.476 | 0.2660 | Yes |
| 21 | Gpr183 | 1847 | 0.470 | 0.2757 | Yes |
| 22 | Gadd45a | 1875 | 0.466 | 0.2867 | Yes |
| 23 | Jun | 2116 | 0.428 | 0.2780 | No |
| 24 | Map3k8 | 2201 | 0.414 | 0.2826 | No |
| 25 | Sphk1 | 2314 | 0.402 | 0.2844 | No |
| 26 | Mxd1 | 2608 | 0.365 | 0.2693 | No |
| 27 | Smad3 | 2656 | 0.359 | 0.2755 | No |
| 28 | Ldlr | 2692 | 0.355 | 0.2826 | No |
| 29 | Ripk2 | 3002 | 0.324 | 0.2650 | No |
| 30 | Ier5 | 3005 | 0.323 | 0.2741 | No |
| 31 | Dusp1 | 3280 | 0.298 | 0.2588 | No |
| 32 | Nfat5 | 3346 | 0.290 | 0.2615 | No |
| 33 | Ptger4 | 3442 | 0.280 | 0.2612 | No |
| 34 | Sqstm1 | 3498 | 0.273 | 0.2643 | No |
| 35 | Pde4b | 3530 | 0.270 | 0.2693 | No |
| 36 | Yrdc | 3632 | 0.260 | 0.2679 | No |
| 37 | Cdkn1a | 4009 | 0.220 | 0.2415 | No |
| 38 | Irs2 | 4170 | 0.202 | 0.2334 | No |
| 39 | Map2k3 | 4255 | 0.194 | 0.2316 | No |
| 40 | Pdlim5 | 4364 | 0.183 | 0.2275 | No |
| 41 | Btg2 | 4373 | 0.183 | 0.2320 | No |
| 42 | Zbtb10 | 4443 | 0.179 | 0.2311 | No |
| 43 | Nr4a2 | 4508 | 0.174 | 0.2305 | No |
| 44 | Cd44 | 4660 | 0.158 | 0.2219 | No |
| 45 | Kdm6b | 4699 | 0.154 | 0.2230 | No |
| 46 | Icam1 | 4710 | 0.153 | 0.2266 | No |
| 47 | Marcks | 4772 | 0.149 | 0.2255 | No |
| 48 | Slc16a6 | 4802 | 0.147 | 0.2272 | No |
| 49 | Slc2a3 | 5009 | 0.130 | 0.2130 | No |
| 50 | Csf1 | 5255 | 0.108 | 0.1948 | No |
| 51 | Trib1 | 5465 | 0.089 | 0.1792 | No |
| 52 | Ppp1r15a | 5467 | 0.089 | 0.1816 | No |
| 53 | Klf10 | 5519 | 0.084 | 0.1796 | No |
| 54 | Tnip2 | 5535 | 0.083 | 0.1807 | No |
| 55 | Tiparp | 5629 | 0.074 | 0.1747 | No |
| 56 | Ptpre | 5996 | 0.043 | 0.1441 | No |
| 57 | Vegfa | 6069 | 0.038 | 0.1389 | No |
| 58 | Birc3 | 6140 | 0.033 | 0.1337 | No |
| 59 | Nfkbia | 6151 | 0.032 | 0.1338 | No |
| 60 | Fosb | 6182 | 0.030 | 0.1320 | No |
| 61 | Sat1 | 6220 | 0.026 | 0.1296 | No |
| 62 | Rela | 6223 | 0.026 | 0.1301 | No |
| 63 | Dram1 | 6256 | 0.023 | 0.1280 | No |
| 64 | Bcl3 | 6551 | 0.000 | 0.1024 | No |
| 65 | Gadd45b | 6647 | -0.004 | 0.0943 | No |
| 66 | Plek | 6749 | -0.012 | 0.0858 | No |
| 67 | Clcf1 | 6873 | -0.024 | 0.0758 | No |
| 68 | Cflar | 6912 | -0.028 | 0.0733 | No |
| 69 | Dusp2 | 7054 | -0.039 | 0.0621 | No |
| 70 | Il15ra | 7057 | -0.039 | 0.0631 | No |
| 71 | Rhob | 7101 | -0.042 | 0.0606 | No |
| 72 | Panx1 | 7182 | -0.050 | 0.0550 | No |
| 73 | Ifih1 | 7206 | -0.052 | 0.0545 | No |
| 74 | Tgif1 | 7417 | -0.069 | 0.0382 | No |
| 75 | Olr1 | 7450 | -0.071 | 0.0375 | No |
| 76 | Klf6 | 7518 | -0.077 | 0.0338 | No |
| 77 | Rel | 7675 | -0.089 | 0.0228 | No |
| 78 | Ier2 | 7777 | -0.096 | 0.0168 | No |
| 79 | Abca1 | 7948 | -0.110 | 0.0051 | No |
| 80 | Tank | 8006 | -0.115 | 0.0035 | No |
| 81 | Myc | 8109 | -0.124 | -0.0019 | No |
| 82 | Nfe2l2 | 8221 | -0.135 | -0.0076 | No |
| 83 | Fos | 8302 | -0.141 | -0.0106 | No |
| 84 | Ets2 | 8360 | -0.146 | -0.0114 | No |
| 85 | Sgk1 | 8520 | -0.159 | -0.0206 | No |
| 86 | Litaf | 8531 | -0.160 | -0.0169 | No |
| 87 | Tnip1 | 8767 | -0.180 | -0.0322 | No |
| 88 | Rigi | 8838 | -0.186 | -0.0330 | No |
| 89 | Pnrc1 | 8901 | -0.191 | -0.0329 | No |
| 90 | Rnf19b | 8904 | -0.191 | -0.0276 | No |
| 91 | Gch1 | 8909 | -0.192 | -0.0225 | No |
| 92 | Tnfaip8 | 9040 | -0.204 | -0.0279 | No |
| 93 | B4galt1 | 9097 | -0.210 | -0.0268 | No |
| 94 | Klf9 | 9116 | -0.211 | -0.0223 | No |
| 95 | Ehd1 | 9172 | -0.216 | -0.0209 | No |
| 96 | Ier3 | 9208 | -0.220 | -0.0176 | No |
| 97 | Ifit2 | 9315 | -0.229 | -0.0203 | No |
| 98 | Dnajb4 | 9344 | -0.232 | -0.0161 | No |
| 99 | Mcl1 | 9494 | -0.247 | -0.0220 | No |
| 100 | B4galt5 | 9524 | -0.250 | -0.0173 | No |
| 101 | Nfkb1 | 9709 | -0.267 | -0.0257 | No |
| 102 | Cebpb | 9748 | -0.270 | -0.0212 | No |
| 103 | Atp2b1 | 9999 | -0.295 | -0.0345 | No |
| 104 | Ccrl2 | 10024 | -0.297 | -0.0281 | No |
| 105 | Nampt | 10273 | -0.324 | -0.0404 | No |
| 106 | Per1 | 10293 | -0.326 | -0.0327 | No |
| 107 | Nfkb2 | 10420 | -0.342 | -0.0339 | No |
| 108 | Birc2 | 10680 | -0.382 | -0.0455 | No |
| 109 | Tsc22d1 | 10687 | -0.383 | -0.0350 | No |
| 110 | Btg1 | 10736 | -0.388 | -0.0281 | No |
| 111 | Eif1 | 10746 | -0.389 | -0.0177 | No |
| 112 | Tnfaip2 | 10795 | -0.398 | -0.0105 | No |
| 113 | Stat5a | 10826 | -0.403 | -0.0016 | No |
| 114 | Rcan1 | 10917 | -0.423 | 0.0027 | No |
| 115 | Lamb3 | 10922 | -0.425 | 0.0146 | No |
| 116 | Fosl2 | 10966 | -0.433 | 0.0232 | No |
| 117 | Ccnl1 | 11342 | -0.561 | 0.0067 | No |
| 118 | Pfkfb3 | 11379 | -0.576 | 0.0200 | No |