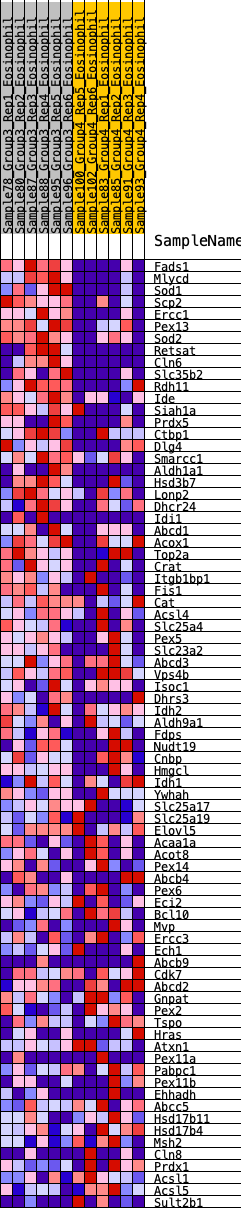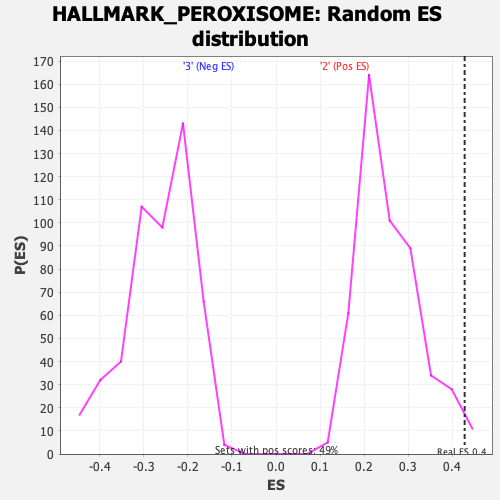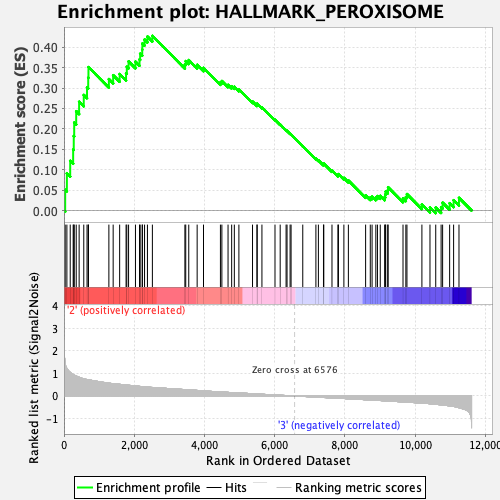
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group3_versus_Group4.Eosinophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.42771068 |
| Normalized Enrichment Score (NES) | 1.6689464 |
| Nominal p-value | 0.020283977 |
| FDR q-value | 0.06660256 |
| FWER p-Value | 0.098 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fads1 | 34 | 1.458 | 0.0507 | Yes |
| 2 | Mlycd | 82 | 1.216 | 0.0914 | Yes |
| 3 | Sod1 | 177 | 1.046 | 0.1218 | Yes |
| 4 | Scp2 | 261 | 0.950 | 0.1495 | Yes |
| 5 | Ercc1 | 278 | 0.936 | 0.1826 | Yes |
| 6 | Pex13 | 291 | 0.923 | 0.2155 | Yes |
| 7 | Sod2 | 348 | 0.880 | 0.2431 | Yes |
| 8 | Retsat | 431 | 0.827 | 0.2664 | Yes |
| 9 | Cln6 | 563 | 0.759 | 0.2830 | Yes |
| 10 | Slc35b2 | 656 | 0.726 | 0.3018 | Yes |
| 11 | Rdh11 | 690 | 0.719 | 0.3254 | Yes |
| 12 | Ide | 697 | 0.717 | 0.3513 | Yes |
| 13 | Siah1a | 1278 | 0.568 | 0.3219 | Yes |
| 14 | Prdx5 | 1400 | 0.542 | 0.3313 | Yes |
| 15 | Ctbp1 | 1585 | 0.515 | 0.3343 | Yes |
| 16 | Dlg4 | 1768 | 0.481 | 0.3363 | Yes |
| 17 | Smarcc1 | 1789 | 0.478 | 0.3521 | Yes |
| 18 | Aldh1a1 | 1839 | 0.471 | 0.3652 | Yes |
| 19 | Hsd3b7 | 2035 | 0.441 | 0.3646 | Yes |
| 20 | Lonp2 | 2151 | 0.422 | 0.3701 | Yes |
| 21 | Dhcr24 | 2169 | 0.420 | 0.3841 | Yes |
| 22 | Idi1 | 2228 | 0.411 | 0.3942 | Yes |
| 23 | Abcd1 | 2232 | 0.411 | 0.4091 | Yes |
| 24 | Acox1 | 2294 | 0.405 | 0.4187 | Yes |
| 25 | Top2a | 2376 | 0.390 | 0.4260 | Yes |
| 26 | Crat | 2517 | 0.375 | 0.4277 | Yes |
| 27 | Itgb1bp1 | 3443 | 0.280 | 0.3578 | No |
| 28 | Fis1 | 3467 | 0.277 | 0.3660 | No |
| 29 | Cat | 3553 | 0.267 | 0.3685 | No |
| 30 | Acsl4 | 3792 | 0.244 | 0.3568 | No |
| 31 | Slc25a4 | 3973 | 0.222 | 0.3493 | No |
| 32 | Pex5 | 4455 | 0.178 | 0.3142 | No |
| 33 | Slc23a2 | 4496 | 0.175 | 0.3171 | No |
| 34 | Abcd3 | 4672 | 0.157 | 0.3077 | No |
| 35 | Vps4b | 4775 | 0.149 | 0.3044 | No |
| 36 | Isoc1 | 4851 | 0.142 | 0.3031 | No |
| 37 | Dhrs3 | 4981 | 0.132 | 0.2968 | No |
| 38 | Idh2 | 5369 | 0.099 | 0.2669 | No |
| 39 | Aldh9a1 | 5497 | 0.086 | 0.2590 | No |
| 40 | Fdps | 5500 | 0.086 | 0.2620 | No |
| 41 | Nudt19 | 5637 | 0.073 | 0.2529 | No |
| 42 | Cnbp | 6009 | 0.042 | 0.2223 | No |
| 43 | Hmgcl | 6157 | 0.032 | 0.2107 | No |
| 44 | Idh1 | 6325 | 0.017 | 0.1968 | No |
| 45 | Ywhah | 6352 | 0.015 | 0.1951 | No |
| 46 | Slc25a17 | 6439 | 0.008 | 0.1880 | No |
| 47 | Slc25a19 | 6467 | 0.006 | 0.1859 | No |
| 48 | Elovl5 | 6800 | -0.016 | 0.1577 | No |
| 49 | Acaa1a | 7174 | -0.049 | 0.1271 | No |
| 50 | Acot8 | 7245 | -0.056 | 0.1231 | No |
| 51 | Pex14 | 7390 | -0.067 | 0.1131 | No |
| 52 | Abcb4 | 7399 | -0.067 | 0.1149 | No |
| 53 | Pex6 | 7632 | -0.084 | 0.0978 | No |
| 54 | Eci2 | 7801 | -0.098 | 0.0869 | No |
| 55 | Bcl10 | 7816 | -0.099 | 0.0893 | No |
| 56 | Mvp | 7971 | -0.111 | 0.0800 | No |
| 57 | Ercc3 | 8099 | -0.124 | 0.0736 | No |
| 58 | Ech1 | 8590 | -0.165 | 0.0372 | No |
| 59 | Abcb9 | 8723 | -0.176 | 0.0322 | No |
| 60 | Cdk7 | 8774 | -0.180 | 0.0345 | No |
| 61 | Abcd2 | 8881 | -0.189 | 0.0323 | No |
| 62 | Gnpat | 8927 | -0.193 | 0.0355 | No |
| 63 | Pex2 | 9007 | -0.201 | 0.0360 | No |
| 64 | Tspo | 9133 | -0.213 | 0.0330 | No |
| 65 | Hras | 9150 | -0.214 | 0.0395 | No |
| 66 | Atxn1 | 9162 | -0.216 | 0.0465 | No |
| 67 | Pex11a | 9220 | -0.221 | 0.0497 | No |
| 68 | Pabpc1 | 9230 | -0.222 | 0.0571 | No |
| 69 | Pex11b | 9655 | -0.263 | 0.0300 | No |
| 70 | Ehhadh | 9736 | -0.270 | 0.0330 | No |
| 71 | Abcc5 | 9767 | -0.272 | 0.0404 | No |
| 72 | Hsd17b11 | 10193 | -0.314 | 0.0151 | No |
| 73 | Hsd17b4 | 10423 | -0.342 | 0.0078 | No |
| 74 | Msh2 | 10587 | -0.367 | 0.0072 | No |
| 75 | Cln8 | 10740 | -0.389 | 0.0083 | No |
| 76 | Prdx1 | 10783 | -0.396 | 0.0193 | No |
| 77 | Acsl1 | 10983 | -0.435 | 0.0180 | No |
| 78 | Acsl5 | 11096 | -0.462 | 0.0253 | No |
| 79 | Sult2b1 | 11249 | -0.518 | 0.0312 | No |