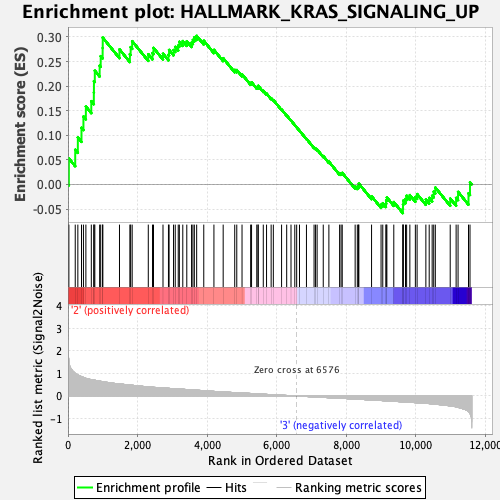
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group3_versus_Group4.Eosinophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_UP |
| Enrichment Score (ES) | 0.3022152 |
| Normalized Enrichment Score (NES) | 1.3177122 |
| Nominal p-value | 0.08461539 |
| FDR q-value | 0.22948343 |
| FWER p-Value | 0.616 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rbm4 | 25 | 1.633 | 0.0529 | Yes |
| 2 | Irf8 | 208 | 0.996 | 0.0706 | Yes |
| 3 | Ccnd2 | 283 | 0.931 | 0.0956 | Yes |
| 4 | Etv5 | 388 | 0.853 | 0.1153 | Yes |
| 5 | Mtmr10 | 444 | 0.820 | 0.1382 | Yes |
| 6 | Tnfaip3 | 514 | 0.779 | 0.1584 | Yes |
| 7 | Plaur | 670 | 0.723 | 0.1694 | Yes |
| 8 | Ank | 743 | 0.701 | 0.1867 | Yes |
| 9 | Tmem176b | 746 | 0.700 | 0.2102 | Yes |
| 10 | F13a1 | 771 | 0.692 | 0.2314 | Yes |
| 11 | H2bc3 | 910 | 0.658 | 0.2416 | Yes |
| 12 | Il1rl2 | 940 | 0.648 | 0.2609 | Yes |
| 13 | Dusp6 | 996 | 0.632 | 0.2775 | Yes |
| 14 | Hsd11b1 | 999 | 0.631 | 0.2986 | Yes |
| 15 | Nrp1 | 1480 | 0.530 | 0.2748 | Yes |
| 16 | Spry2 | 1779 | 0.480 | 0.2651 | Yes |
| 17 | Zfp639 | 1801 | 0.476 | 0.2793 | Yes |
| 18 | Eng | 1845 | 0.470 | 0.2914 | Yes |
| 19 | Itgb2 | 2308 | 0.403 | 0.2648 | Yes |
| 20 | Avl9 | 2429 | 0.384 | 0.2673 | Yes |
| 21 | Adam17 | 2458 | 0.381 | 0.2778 | Yes |
| 22 | Kcnn4 | 2731 | 0.352 | 0.2660 | Yes |
| 23 | Tmem176a | 2888 | 0.337 | 0.2638 | Yes |
| 24 | Mmd | 2906 | 0.334 | 0.2736 | Yes |
| 25 | Lat2 | 3034 | 0.320 | 0.2734 | Yes |
| 26 | Satb1 | 3083 | 0.315 | 0.2798 | Yes |
| 27 | Jup | 3169 | 0.305 | 0.2827 | Yes |
| 28 | Cbx8 | 3201 | 0.302 | 0.2902 | Yes |
| 29 | Lcp1 | 3300 | 0.296 | 0.2917 | Yes |
| 30 | Angptl4 | 3415 | 0.282 | 0.2913 | Yes |
| 31 | Ikzf1 | 3552 | 0.267 | 0.2885 | Yes |
| 32 | Scg5 | 3591 | 0.264 | 0.2941 | Yes |
| 33 | Yrdc | 3632 | 0.260 | 0.2994 | Yes |
| 34 | Ero1a | 3699 | 0.253 | 0.3022 | Yes |
| 35 | Akt2 | 3903 | 0.229 | 0.2923 | No |
| 36 | Evi5 | 4195 | 0.200 | 0.2738 | No |
| 37 | Plvap | 4461 | 0.177 | 0.2568 | No |
| 38 | Hdac9 | 4793 | 0.147 | 0.2330 | No |
| 39 | Rabgap1l | 4852 | 0.142 | 0.2327 | No |
| 40 | Dock2 | 5003 | 0.131 | 0.2241 | No |
| 41 | Car2 | 5251 | 0.109 | 0.2063 | No |
| 42 | Ptcd2 | 5277 | 0.106 | 0.2077 | No |
| 43 | Ets1 | 5427 | 0.094 | 0.1980 | No |
| 44 | Trib1 | 5465 | 0.089 | 0.1978 | No |
| 45 | Ppp1r15a | 5467 | 0.089 | 0.2007 | No |
| 46 | Ptbp2 | 5614 | 0.076 | 0.1906 | No |
| 47 | Tor1aip2 | 5708 | 0.068 | 0.1848 | No |
| 48 | Map3k1 | 5840 | 0.058 | 0.1754 | No |
| 49 | Laptm5 | 5903 | 0.051 | 0.1717 | No |
| 50 | Birc3 | 6140 | 0.033 | 0.1524 | No |
| 51 | Strn | 6287 | 0.021 | 0.1404 | No |
| 52 | Cdadc1 | 6411 | 0.010 | 0.1300 | No |
| 53 | Tfpi | 6520 | 0.002 | 0.1207 | No |
| 54 | Csf2ra | 6574 | 0.000 | 0.1161 | No |
| 55 | Map7 | 6655 | -0.005 | 0.1094 | No |
| 56 | Zfp277 | 6855 | -0.021 | 0.0928 | No |
| 57 | Cbr4 | 7074 | -0.040 | 0.0752 | No |
| 58 | Dnmbp | 7117 | -0.044 | 0.0730 | No |
| 59 | Cbl | 7162 | -0.049 | 0.0709 | No |
| 60 | Nin | 7338 | -0.062 | 0.0578 | No |
| 61 | Map4k1 | 7500 | -0.076 | 0.0463 | No |
| 62 | Usp12 | 7808 | -0.098 | 0.0230 | No |
| 63 | Galnt3 | 7850 | -0.102 | 0.0229 | No |
| 64 | Il2rg | 7884 | -0.105 | 0.0235 | No |
| 65 | Cxcr4 | 8255 | -0.137 | -0.0040 | No |
| 66 | Gypc | 8318 | -0.143 | -0.0046 | No |
| 67 | Wdr33 | 8353 | -0.145 | -0.0026 | No |
| 68 | Fbxo4 | 8361 | -0.146 | 0.0017 | No |
| 69 | Cab39l | 8727 | -0.176 | -0.0241 | No |
| 70 | Adgrl4 | 9000 | -0.200 | -0.0409 | No |
| 71 | Prelid3b | 9049 | -0.205 | -0.0382 | No |
| 72 | Cd37 | 9140 | -0.214 | -0.0388 | No |
| 73 | Tspan13 | 9148 | -0.214 | -0.0322 | No |
| 74 | Bpgm | 9167 | -0.216 | -0.0265 | No |
| 75 | Cmklr1 | 9366 | -0.233 | -0.0358 | No |
| 76 | Ano1 | 9627 | -0.260 | -0.0496 | No |
| 77 | Crot | 9631 | -0.260 | -0.0411 | No |
| 78 | Btbd3 | 9638 | -0.261 | -0.0328 | No |
| 79 | Tnfrsf1b | 9703 | -0.267 | -0.0294 | No |
| 80 | St6gal1 | 9735 | -0.269 | -0.0230 | No |
| 81 | Itga2 | 9828 | -0.278 | -0.0217 | No |
| 82 | Fcer1g | 9986 | -0.294 | -0.0254 | No |
| 83 | Glrx | 10036 | -0.298 | -0.0196 | No |
| 84 | Atg10 | 10288 | -0.326 | -0.0304 | No |
| 85 | Fuca1 | 10384 | -0.337 | -0.0273 | No |
| 86 | Pdcd1lg2 | 10469 | -0.350 | -0.0228 | No |
| 87 | Adam8 | 10514 | -0.356 | -0.0146 | No |
| 88 | Sdccag8 | 10560 | -0.362 | -0.0063 | No |
| 89 | Vwa5a | 10988 | -0.436 | -0.0287 | No |
| 90 | Ccser2 | 11158 | -0.478 | -0.0273 | No |
| 91 | Ammecr1 | 11214 | -0.503 | -0.0151 | No |
| 92 | Psmb8 | 11514 | -0.684 | -0.0180 | No |
| 93 | Pecam1 | 11557 | -0.777 | 0.0045 | No |

