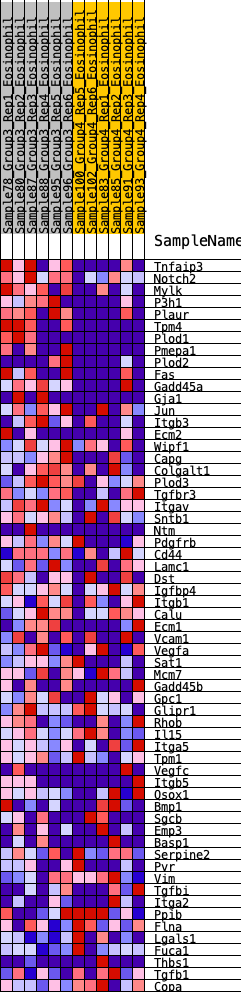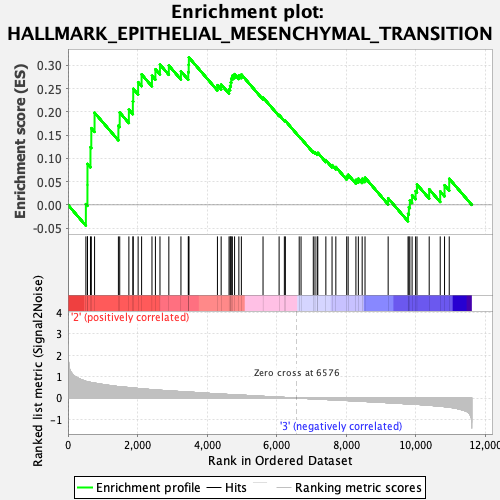
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group3_versus_Group4.Eosinophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.3168257 |
| Normalized Enrichment Score (NES) | 1.3089966 |
| Nominal p-value | 0.053359684 |
| FDR q-value | 0.21447818 |
| FWER p-Value | 0.642 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tnfaip3 | 514 | 0.779 | 0.0017 | Yes |
| 2 | Notch2 | 559 | 0.761 | 0.0430 | Yes |
| 3 | Mylk | 560 | 0.760 | 0.0880 | Yes |
| 4 | P3h1 | 646 | 0.730 | 0.1239 | Yes |
| 5 | Plaur | 670 | 0.723 | 0.1648 | Yes |
| 6 | Tpm4 | 764 | 0.694 | 0.1979 | Yes |
| 7 | Plod1 | 1449 | 0.534 | 0.1703 | Yes |
| 8 | Pmepa1 | 1487 | 0.529 | 0.1985 | Yes |
| 9 | Plod2 | 1750 | 0.487 | 0.2047 | Yes |
| 10 | Fas | 1865 | 0.467 | 0.2225 | Yes |
| 11 | Gadd45a | 1875 | 0.466 | 0.2494 | Yes |
| 12 | Gja1 | 2016 | 0.445 | 0.2636 | Yes |
| 13 | Jun | 2116 | 0.428 | 0.2804 | Yes |
| 14 | Itgb3 | 2414 | 0.386 | 0.2776 | Yes |
| 15 | Ecm2 | 2513 | 0.376 | 0.2913 | Yes |
| 16 | Wipf1 | 2644 | 0.361 | 0.3015 | Yes |
| 17 | Capg | 2899 | 0.335 | 0.2993 | Yes |
| 18 | Colgalt1 | 3247 | 0.300 | 0.2871 | Yes |
| 19 | Plod3 | 3458 | 0.278 | 0.2854 | Yes |
| 20 | Tgfbr3 | 3470 | 0.277 | 0.3008 | Yes |
| 21 | Itgav | 3475 | 0.276 | 0.3168 | Yes |
| 22 | Sntb1 | 4296 | 0.190 | 0.2571 | No |
| 23 | Ntm | 4403 | 0.181 | 0.2586 | No |
| 24 | Pdgfrb | 4630 | 0.161 | 0.2486 | No |
| 25 | Cd44 | 4660 | 0.158 | 0.2555 | No |
| 26 | Lamc1 | 4680 | 0.156 | 0.2631 | No |
| 27 | Dst | 4693 | 0.155 | 0.2713 | No |
| 28 | Igfbp4 | 4725 | 0.152 | 0.2776 | No |
| 29 | Itgb1 | 4789 | 0.148 | 0.2809 | No |
| 30 | Calu | 4912 | 0.136 | 0.2784 | No |
| 31 | Ecm1 | 4984 | 0.132 | 0.2801 | No |
| 32 | Vcam1 | 5607 | 0.077 | 0.2308 | No |
| 33 | Vegfa | 6069 | 0.038 | 0.1931 | No |
| 34 | Sat1 | 6220 | 0.026 | 0.1817 | No |
| 35 | Mcm7 | 6249 | 0.023 | 0.1806 | No |
| 36 | Gadd45b | 6647 | -0.004 | 0.1465 | No |
| 37 | Gpc1 | 6700 | -0.009 | 0.1425 | No |
| 38 | Glipr1 | 7052 | -0.039 | 0.1144 | No |
| 39 | Rhob | 7101 | -0.042 | 0.1127 | No |
| 40 | Il15 | 7163 | -0.049 | 0.1104 | No |
| 41 | Itga5 | 7181 | -0.050 | 0.1119 | No |
| 42 | Tpm1 | 7413 | -0.068 | 0.0959 | No |
| 43 | Vegfc | 7591 | -0.081 | 0.0854 | No |
| 44 | Itgb5 | 7700 | -0.090 | 0.0814 | No |
| 45 | Qsox1 | 8012 | -0.116 | 0.0613 | No |
| 46 | Bmp1 | 8054 | -0.119 | 0.0648 | No |
| 47 | Sgcb | 8277 | -0.139 | 0.0538 | No |
| 48 | Emp3 | 8347 | -0.145 | 0.0564 | No |
| 49 | Basp1 | 8457 | -0.153 | 0.0561 | No |
| 50 | Serpine2 | 8539 | -0.161 | 0.0586 | No |
| 51 | Pvr | 9204 | -0.220 | 0.0141 | No |
| 52 | Vim | 9776 | -0.273 | -0.0191 | No |
| 53 | Tgfbi | 9800 | -0.275 | -0.0048 | No |
| 54 | Itga2 | 9828 | -0.278 | 0.0093 | No |
| 55 | Ppib | 9892 | -0.285 | 0.0208 | No |
| 56 | Flna | 9993 | -0.294 | 0.0296 | No |
| 57 | Lgals1 | 10033 | -0.298 | 0.0439 | No |
| 58 | Fuca1 | 10384 | -0.337 | 0.0335 | No |
| 59 | Thbs1 | 10701 | -0.384 | 0.0289 | No |
| 60 | Tgfb1 | 10825 | -0.403 | 0.0422 | No |
| 61 | Copa | 10960 | -0.432 | 0.0562 | No |