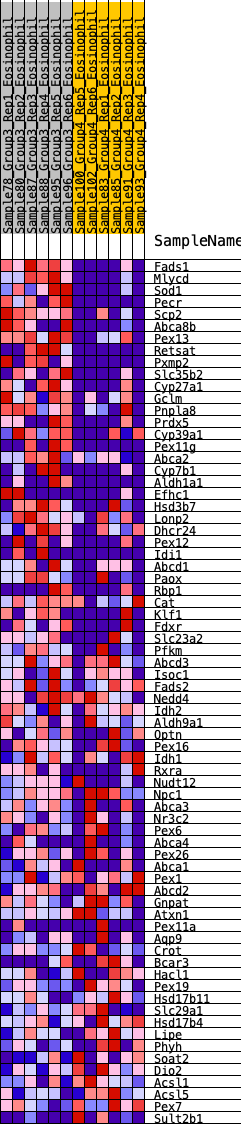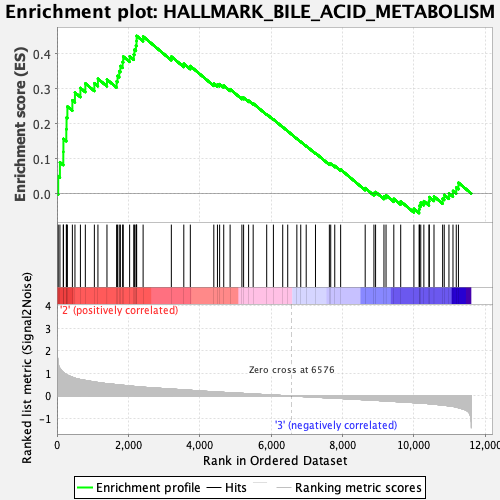
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group3_versus_Group4.Eosinophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.45191237 |
| Normalized Enrichment Score (NES) | 1.694978 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07756086 |
| FWER p-Value | 0.084 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fads1 | 34 | 1.458 | 0.0498 | Yes |
| 2 | Mlycd | 82 | 1.216 | 0.0898 | Yes |
| 3 | Sod1 | 177 | 1.046 | 0.1195 | Yes |
| 4 | Pecr | 180 | 1.038 | 0.1569 | Yes |
| 5 | Scp2 | 261 | 0.950 | 0.1843 | Yes |
| 6 | Abca8b | 268 | 0.945 | 0.2180 | Yes |
| 7 | Pex13 | 291 | 0.923 | 0.2495 | Yes |
| 8 | Retsat | 431 | 0.827 | 0.2674 | Yes |
| 9 | Pxmp2 | 503 | 0.785 | 0.2896 | Yes |
| 10 | Slc35b2 | 656 | 0.726 | 0.3027 | Yes |
| 11 | Cyp27a1 | 794 | 0.686 | 0.3157 | Yes |
| 12 | Gclm | 1047 | 0.616 | 0.3161 | Yes |
| 13 | Pnpla8 | 1147 | 0.596 | 0.3291 | Yes |
| 14 | Prdx5 | 1400 | 0.542 | 0.3269 | Yes |
| 15 | Cyp39a1 | 1670 | 0.499 | 0.3216 | Yes |
| 16 | Pex11g | 1697 | 0.495 | 0.3373 | Yes |
| 17 | Abca2 | 1744 | 0.487 | 0.3509 | Yes |
| 18 | Cyp7b1 | 1776 | 0.480 | 0.3656 | Yes |
| 19 | Aldh1a1 | 1839 | 0.471 | 0.3773 | Yes |
| 20 | Efhc1 | 1856 | 0.469 | 0.3928 | Yes |
| 21 | Hsd3b7 | 2035 | 0.441 | 0.3934 | Yes |
| 22 | Lonp2 | 2151 | 0.422 | 0.3987 | Yes |
| 23 | Dhcr24 | 2169 | 0.420 | 0.4124 | Yes |
| 24 | Pex12 | 2212 | 0.413 | 0.4237 | Yes |
| 25 | Idi1 | 2228 | 0.411 | 0.4373 | Yes |
| 26 | Abcd1 | 2232 | 0.411 | 0.4519 | Yes |
| 27 | Paox | 2413 | 0.386 | 0.4503 | No |
| 28 | Rbp1 | 3204 | 0.302 | 0.3928 | No |
| 29 | Cat | 3553 | 0.267 | 0.3723 | No |
| 30 | Klf1 | 3736 | 0.250 | 0.3655 | No |
| 31 | Fdxr | 4394 | 0.181 | 0.3151 | No |
| 32 | Slc23a2 | 4496 | 0.175 | 0.3127 | No |
| 33 | Pfkm | 4557 | 0.168 | 0.3136 | No |
| 34 | Abcd3 | 4672 | 0.157 | 0.3094 | No |
| 35 | Isoc1 | 4851 | 0.142 | 0.2991 | No |
| 36 | Fads2 | 5181 | 0.115 | 0.2748 | No |
| 37 | Nedd4 | 5229 | 0.111 | 0.2747 | No |
| 38 | Idh2 | 5369 | 0.099 | 0.2662 | No |
| 39 | Aldh9a1 | 5497 | 0.086 | 0.2583 | No |
| 40 | Optn | 5876 | 0.054 | 0.2275 | No |
| 41 | Pex16 | 6064 | 0.038 | 0.2127 | No |
| 42 | Idh1 | 6325 | 0.017 | 0.1908 | No |
| 43 | Rxra | 6465 | 0.007 | 0.1790 | No |
| 44 | Nudt12 | 6721 | -0.010 | 0.1572 | No |
| 45 | Npc1 | 6830 | -0.019 | 0.1486 | No |
| 46 | Abca3 | 6983 | -0.034 | 0.1366 | No |
| 47 | Nr3c2 | 7243 | -0.056 | 0.1162 | No |
| 48 | Pex6 | 7632 | -0.084 | 0.0856 | No |
| 49 | Abca4 | 7670 | -0.088 | 0.0856 | No |
| 50 | Pex26 | 7783 | -0.097 | 0.0794 | No |
| 51 | Abca1 | 7948 | -0.110 | 0.0691 | No |
| 52 | Pex1 | 8636 | -0.169 | 0.0157 | No |
| 53 | Abcd2 | 8881 | -0.189 | 0.0014 | No |
| 54 | Gnpat | 8927 | -0.193 | 0.0045 | No |
| 55 | Atxn1 | 9162 | -0.216 | -0.0080 | No |
| 56 | Pex11a | 9220 | -0.221 | -0.0049 | No |
| 57 | Aqp9 | 9438 | -0.242 | -0.0150 | No |
| 58 | Crot | 9631 | -0.260 | -0.0222 | No |
| 59 | Bcar3 | 10001 | -0.295 | -0.0435 | No |
| 60 | Hacl1 | 10147 | -0.310 | -0.0449 | No |
| 61 | Pex19 | 10158 | -0.311 | -0.0345 | No |
| 62 | Hsd17b11 | 10193 | -0.314 | -0.0261 | No |
| 63 | Slc29a1 | 10280 | -0.326 | -0.0218 | No |
| 64 | Hsd17b4 | 10423 | -0.342 | -0.0217 | No |
| 65 | Lipe | 10433 | -0.344 | -0.0100 | No |
| 66 | Phyh | 10564 | -0.363 | -0.0082 | No |
| 67 | Soat2 | 10806 | -0.399 | -0.0146 | No |
| 68 | Dio2 | 10851 | -0.408 | -0.0037 | No |
| 69 | Acsl1 | 10983 | -0.435 | 0.0007 | No |
| 70 | Acsl5 | 11096 | -0.462 | 0.0077 | No |
| 71 | Pex7 | 11188 | -0.491 | 0.0176 | No |
| 72 | Sult2b1 | 11249 | -0.518 | 0.0312 | No |