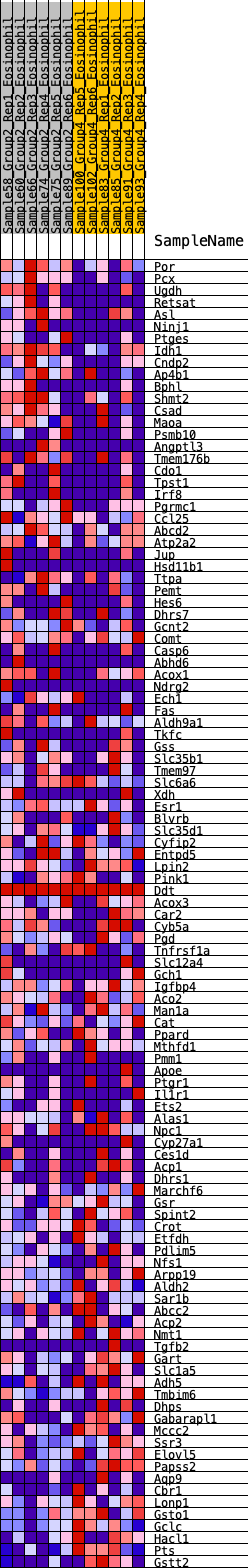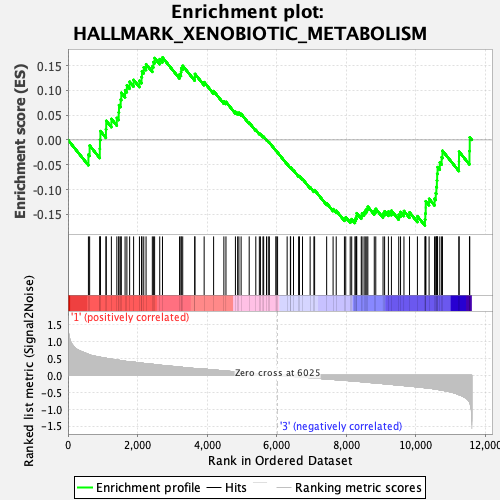
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group4.Eosinophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_XENOBIOTIC_METABOLISM |
| Enrichment Score (ES) | -0.17264377 |
| Normalized Enrichment Score (NES) | -0.75707877 |
| Nominal p-value | 0.9158317 |
| FDR q-value | 0.8650386 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Por | 587 | 0.621 | -0.0295 | Yes |
| 2 | Pcx | 622 | 0.608 | -0.0113 | Yes |
| 3 | Ugdh | 914 | 0.537 | -0.0179 | Yes |
| 4 | Retsat | 919 | 0.537 | 0.0003 | Yes |
| 5 | Asl | 932 | 0.533 | 0.0178 | Yes |
| 6 | Ninj1 | 1091 | 0.505 | 0.0216 | Yes |
| 7 | Ptges | 1099 | 0.504 | 0.0385 | Yes |
| 8 | Idh1 | 1247 | 0.479 | 0.0424 | Yes |
| 9 | Cndp2 | 1404 | 0.453 | 0.0445 | Yes |
| 10 | Ap4b1 | 1458 | 0.447 | 0.0554 | Yes |
| 11 | Bphl | 1466 | 0.445 | 0.0703 | Yes |
| 12 | Shmt2 | 1515 | 0.439 | 0.0814 | Yes |
| 13 | Csad | 1532 | 0.437 | 0.0951 | Yes |
| 14 | Maoa | 1641 | 0.419 | 0.1003 | Yes |
| 15 | Psmb10 | 1693 | 0.409 | 0.1101 | Yes |
| 16 | Angptl3 | 1772 | 0.403 | 0.1173 | Yes |
| 17 | Tmem176b | 1886 | 0.389 | 0.1210 | Yes |
| 18 | Cdo1 | 2054 | 0.370 | 0.1193 | Yes |
| 19 | Tpst1 | 2114 | 0.363 | 0.1268 | Yes |
| 20 | Irf8 | 2126 | 0.361 | 0.1384 | Yes |
| 21 | Pgrmc1 | 2178 | 0.353 | 0.1462 | Yes |
| 22 | Ccl25 | 2244 | 0.346 | 0.1526 | Yes |
| 23 | Abcd2 | 2424 | 0.327 | 0.1484 | Yes |
| 24 | Atp2a2 | 2455 | 0.324 | 0.1570 | Yes |
| 25 | Jup | 2490 | 0.320 | 0.1651 | Yes |
| 26 | Hsd11b1 | 2637 | 0.301 | 0.1629 | Yes |
| 27 | Ttpa | 2716 | 0.293 | 0.1663 | Yes |
| 28 | Pemt | 3207 | 0.245 | 0.1322 | No |
| 29 | Hes6 | 3249 | 0.241 | 0.1370 | No |
| 30 | Dhrs7 | 3255 | 0.240 | 0.1449 | No |
| 31 | Gcnt2 | 3298 | 0.235 | 0.1494 | No |
| 32 | Comt | 3643 | 0.202 | 0.1265 | No |
| 33 | Casp6 | 3649 | 0.201 | 0.1331 | No |
| 34 | Abhd6 | 3914 | 0.184 | 0.1165 | No |
| 35 | Acox1 | 4186 | 0.159 | 0.0985 | No |
| 36 | Ndrg2 | 4481 | 0.130 | 0.0774 | No |
| 37 | Ech1 | 4541 | 0.128 | 0.0767 | No |
| 38 | Fas | 4813 | 0.102 | 0.0567 | No |
| 39 | Aldh9a1 | 4877 | 0.096 | 0.0546 | No |
| 40 | Tkfc | 4913 | 0.093 | 0.0548 | No |
| 41 | Gss | 4977 | 0.090 | 0.0524 | No |
| 42 | Slc35b1 | 5209 | 0.069 | 0.0347 | No |
| 43 | Tmem97 | 5399 | 0.055 | 0.0202 | No |
| 44 | Slc6a6 | 5501 | 0.045 | 0.0130 | No |
| 45 | Xdh | 5532 | 0.043 | 0.0119 | No |
| 46 | Esr1 | 5605 | 0.035 | 0.0068 | No |
| 47 | Blvrb | 5625 | 0.033 | 0.0063 | No |
| 48 | Slc35d1 | 5707 | 0.026 | 0.0002 | No |
| 49 | Cyfip2 | 5757 | 0.023 | -0.0033 | No |
| 50 | Entpd5 | 5789 | 0.019 | -0.0053 | No |
| 51 | Lpin2 | 5980 | 0.004 | -0.0217 | No |
| 52 | Pink1 | 5981 | 0.004 | -0.0216 | No |
| 53 | Ddt | 6025 | 0.000 | -0.0253 | No |
| 54 | Acox3 | 6300 | -0.017 | -0.0486 | No |
| 55 | Car2 | 6393 | -0.025 | -0.0557 | No |
| 56 | Cyb5a | 6400 | -0.026 | -0.0553 | No |
| 57 | Pgd | 6483 | -0.032 | -0.0614 | No |
| 58 | Tnfrsf1a | 6630 | -0.044 | -0.0725 | No |
| 59 | Slc12a4 | 6654 | -0.046 | -0.0729 | No |
| 60 | Gch1 | 6742 | -0.050 | -0.0787 | No |
| 61 | Igfbp4 | 6961 | -0.067 | -0.0953 | No |
| 62 | Aco2 | 7070 | -0.075 | -0.1021 | No |
| 63 | Man1a | 7090 | -0.076 | -0.1011 | No |
| 64 | Cat | 7436 | -0.104 | -0.1275 | No |
| 65 | Ppard | 7621 | -0.120 | -0.1394 | No |
| 66 | Mthfd1 | 7711 | -0.127 | -0.1427 | No |
| 67 | Pmm1 | 7947 | -0.148 | -0.1580 | No |
| 68 | Apoe | 7985 | -0.151 | -0.1560 | No |
| 69 | Ptgr1 | 8113 | -0.158 | -0.1615 | No |
| 70 | Il1r1 | 8156 | -0.162 | -0.1595 | No |
| 71 | Ets2 | 8249 | -0.170 | -0.1616 | No |
| 72 | Alas1 | 8273 | -0.172 | -0.1576 | No |
| 73 | Npc1 | 8296 | -0.175 | -0.1535 | No |
| 74 | Cyp27a1 | 8300 | -0.176 | -0.1476 | No |
| 75 | Ces1d | 8431 | -0.184 | -0.1525 | No |
| 76 | Acp1 | 8453 | -0.187 | -0.1479 | No |
| 77 | Dhrs1 | 8511 | -0.193 | -0.1461 | No |
| 78 | Marchf6 | 8546 | -0.197 | -0.1423 | No |
| 79 | Gsr | 8589 | -0.200 | -0.1390 | No |
| 80 | Spint2 | 8620 | -0.203 | -0.1345 | No |
| 81 | Crot | 8805 | -0.219 | -0.1429 | No |
| 82 | Etfdh | 8848 | -0.222 | -0.1388 | No |
| 83 | Pdlim5 | 9057 | -0.243 | -0.1485 | No |
| 84 | Nfs1 | 9102 | -0.247 | -0.1438 | No |
| 85 | Arpp19 | 9211 | -0.256 | -0.1442 | No |
| 86 | Aldh2 | 9298 | -0.266 | -0.1425 | No |
| 87 | Sar1b | 9508 | -0.286 | -0.1507 | No |
| 88 | Abcc2 | 9560 | -0.289 | -0.1451 | No |
| 89 | Acp2 | 9658 | -0.300 | -0.1431 | No |
| 90 | Nmt1 | 9818 | -0.318 | -0.1459 | No |
| 91 | Tgfb2 | 10045 | -0.337 | -0.1539 | No |
| 92 | Gart | 10262 | -0.365 | -0.1600 | No |
| 93 | Slc1a5 | 10273 | -0.366 | -0.1481 | No |
| 94 | Adh5 | 10284 | -0.367 | -0.1362 | No |
| 95 | Tmbim6 | 10285 | -0.367 | -0.1235 | No |
| 96 | Dhps | 10381 | -0.379 | -0.1186 | No |
| 97 | Gabarapl1 | 10538 | -0.398 | -0.1183 | No |
| 98 | Mccc2 | 10573 | -0.402 | -0.1073 | No |
| 99 | Ssr3 | 10590 | -0.406 | -0.0946 | No |
| 100 | Elovl5 | 10605 | -0.408 | -0.0817 | No |
| 101 | Papss2 | 10613 | -0.409 | -0.0681 | No |
| 102 | Aqp9 | 10625 | -0.412 | -0.0548 | No |
| 103 | Cbr1 | 10689 | -0.424 | -0.0455 | No |
| 104 | Lonp1 | 10741 | -0.432 | -0.0349 | No |
| 105 | Gsto1 | 10765 | -0.436 | -0.0218 | No |
| 106 | Gclc | 11239 | -0.570 | -0.0431 | No |
| 107 | Hacl1 | 11242 | -0.570 | -0.0235 | No |
| 108 | Pts | 11542 | -0.783 | -0.0223 | No |
| 109 | Gstt2 | 11550 | -0.807 | 0.0051 | No |