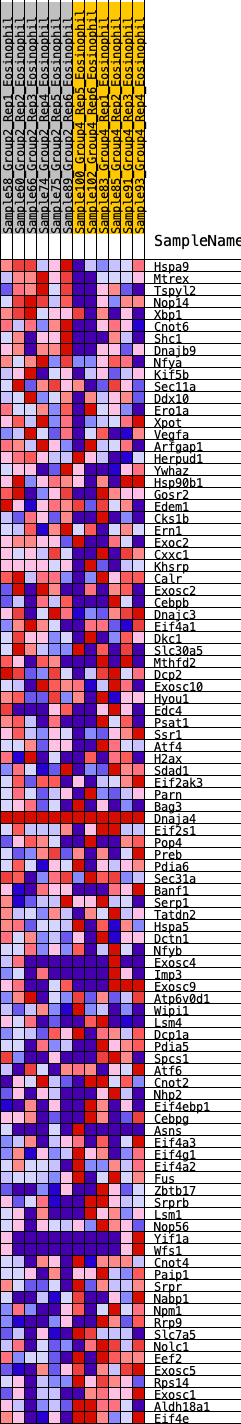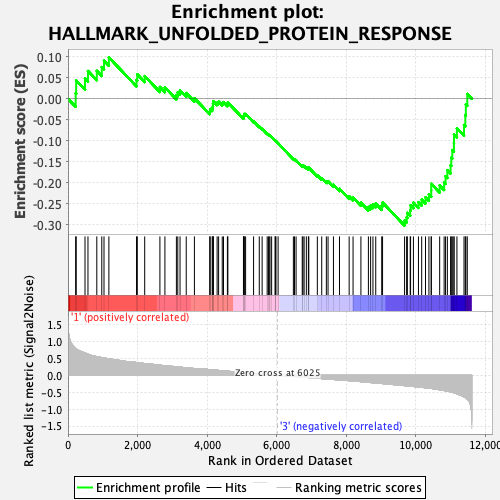
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group4.Eosinophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | -0.30175203 |
| Normalized Enrichment Score (NES) | -1.2112763 |
| Nominal p-value | 0.19838056 |
| FDR q-value | 0.3659557 |
| FWER p-Value | 0.849 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hspa9 | 220 | 0.797 | 0.0130 | No |
| 2 | Mtrex | 236 | 0.787 | 0.0434 | No |
| 3 | Tspyl2 | 489 | 0.659 | 0.0481 | No |
| 4 | Nop14 | 574 | 0.625 | 0.0660 | No |
| 5 | Xbp1 | 828 | 0.556 | 0.0664 | No |
| 6 | Cnot6 | 971 | 0.527 | 0.0753 | No |
| 7 | Shc1 | 1035 | 0.515 | 0.0906 | No |
| 8 | Dnajb9 | 1175 | 0.491 | 0.0983 | No |
| 9 | Nfya | 1970 | 0.380 | 0.0447 | No |
| 10 | Kif5b | 1992 | 0.378 | 0.0581 | No |
| 11 | Sec11a | 2205 | 0.351 | 0.0538 | No |
| 12 | Ddx10 | 2642 | 0.300 | 0.0280 | No |
| 13 | Ero1a | 2786 | 0.285 | 0.0271 | No |
| 14 | Xpot | 3116 | 0.255 | 0.0088 | No |
| 15 | Vegfa | 3156 | 0.251 | 0.0155 | No |
| 16 | Arfgap1 | 3221 | 0.244 | 0.0198 | No |
| 17 | Herpud1 | 3399 | 0.224 | 0.0135 | No |
| 18 | Ywhaz | 3634 | 0.202 | 0.0013 | No |
| 19 | Hsp90b1 | 4074 | 0.169 | -0.0300 | No |
| 20 | Gosr2 | 4088 | 0.168 | -0.0244 | No |
| 21 | Edem1 | 4145 | 0.163 | -0.0227 | No |
| 22 | Cks1b | 4164 | 0.162 | -0.0177 | No |
| 23 | Ern1 | 4169 | 0.161 | -0.0115 | No |
| 24 | Exoc2 | 4178 | 0.161 | -0.0058 | No |
| 25 | Cxxc1 | 4287 | 0.151 | -0.0091 | No |
| 26 | Khsrp | 4330 | 0.146 | -0.0068 | No |
| 27 | Calr | 4434 | 0.135 | -0.0104 | No |
| 28 | Exosc2 | 4470 | 0.131 | -0.0081 | No |
| 29 | Cebpb | 4582 | 0.124 | -0.0128 | No |
| 30 | Dnajc3 | 4596 | 0.123 | -0.0089 | No |
| 31 | Eif4a1 | 5044 | 0.084 | -0.0444 | No |
| 32 | Dkc1 | 5048 | 0.083 | -0.0413 | No |
| 33 | Slc30a5 | 5062 | 0.082 | -0.0391 | No |
| 34 | Mthfd2 | 5067 | 0.082 | -0.0362 | No |
| 35 | Dcp2 | 5106 | 0.078 | -0.0363 | No |
| 36 | Exosc10 | 5335 | 0.061 | -0.0537 | No |
| 37 | Hyou1 | 5499 | 0.045 | -0.0660 | No |
| 38 | Edc4 | 5581 | 0.037 | -0.0715 | No |
| 39 | Psat1 | 5726 | 0.024 | -0.0831 | No |
| 40 | Ssr1 | 5762 | 0.022 | -0.0852 | No |
| 41 | Atf4 | 5784 | 0.020 | -0.0863 | No |
| 42 | H2ax | 5807 | 0.018 | -0.0874 | No |
| 43 | Sdad1 | 5854 | 0.014 | -0.0909 | No |
| 44 | Eif2ak3 | 5952 | 0.006 | -0.0991 | No |
| 45 | Parn | 5970 | 0.004 | -0.1004 | No |
| 46 | Bag3 | 5976 | 0.004 | -0.1006 | No |
| 47 | Dnaja4 | 6040 | 0.000 | -0.1061 | No |
| 48 | Eif2s1 | 6479 | -0.032 | -0.1429 | No |
| 49 | Pop4 | 6511 | -0.034 | -0.1442 | No |
| 50 | Preb | 6557 | -0.038 | -0.1466 | No |
| 51 | Pdia6 | 6734 | -0.050 | -0.1598 | No |
| 52 | Sec31a | 6754 | -0.051 | -0.1594 | No |
| 53 | Banf1 | 6788 | -0.053 | -0.1601 | No |
| 54 | Serp1 | 6852 | -0.060 | -0.1632 | No |
| 55 | Tatdn2 | 6911 | -0.064 | -0.1657 | No |
| 56 | Hspa5 | 6923 | -0.064 | -0.1640 | No |
| 57 | Dctn1 | 7168 | -0.083 | -0.1819 | No |
| 58 | Nfyb | 7294 | -0.094 | -0.1889 | No |
| 59 | Exosc4 | 7427 | -0.103 | -0.1962 | No |
| 60 | Imp3 | 7476 | -0.107 | -0.1961 | No |
| 61 | Exosc9 | 7631 | -0.121 | -0.2046 | No |
| 62 | Atp6v0d1 | 7805 | -0.135 | -0.2142 | No |
| 63 | Wipi1 | 8083 | -0.155 | -0.2320 | No |
| 64 | Lsm4 | 8195 | -0.165 | -0.2350 | No |
| 65 | Dcp1a | 8421 | -0.184 | -0.2471 | No |
| 66 | Pdia5 | 8636 | -0.205 | -0.2575 | No |
| 67 | Spcs1 | 8700 | -0.211 | -0.2544 | No |
| 68 | Atf6 | 8767 | -0.216 | -0.2514 | No |
| 69 | Cnot2 | 8847 | -0.222 | -0.2494 | No |
| 70 | Nhp2 | 9019 | -0.239 | -0.2546 | No |
| 71 | Eif4ebp1 | 9043 | -0.241 | -0.2469 | No |
| 72 | Cebpg | 9676 | -0.302 | -0.2896 | Yes |
| 73 | Asns | 9735 | -0.309 | -0.2822 | Yes |
| 74 | Eif4a3 | 9756 | -0.311 | -0.2714 | Yes |
| 75 | Eif4g1 | 9838 | -0.321 | -0.2655 | Yes |
| 76 | Eif4a2 | 9849 | -0.321 | -0.2534 | Yes |
| 77 | Fus | 9930 | -0.328 | -0.2472 | Yes |
| 78 | Zbtb17 | 10075 | -0.340 | -0.2459 | Yes |
| 79 | Srprb | 10168 | -0.351 | -0.2398 | Yes |
| 80 | Lsm1 | 10278 | -0.367 | -0.2345 | Yes |
| 81 | Nop56 | 10376 | -0.378 | -0.2276 | Yes |
| 82 | Yif1a | 10442 | -0.385 | -0.2178 | Yes |
| 83 | Wfs1 | 10444 | -0.385 | -0.2023 | Yes |
| 84 | Cnot4 | 10685 | -0.423 | -0.2061 | Yes |
| 85 | Paip1 | 10813 | -0.447 | -0.1991 | Yes |
| 86 | Srpr | 10854 | -0.456 | -0.1842 | Yes |
| 87 | Nabp1 | 10906 | -0.466 | -0.1699 | Yes |
| 88 | Npm1 | 10999 | -0.488 | -0.1582 | Yes |
| 89 | Rrp9 | 11019 | -0.492 | -0.1400 | Yes |
| 90 | Slc7a5 | 11048 | -0.500 | -0.1223 | Yes |
| 91 | Nolc1 | 11095 | -0.514 | -0.1056 | Yes |
| 92 | Eef2 | 11098 | -0.515 | -0.0850 | Yes |
| 93 | Exosc5 | 11181 | -0.543 | -0.0703 | Yes |
| 94 | Rps14 | 11385 | -0.632 | -0.0624 | Yes |
| 95 | Exosc1 | 11423 | -0.659 | -0.0391 | Yes |
| 96 | Aldh18a1 | 11438 | -0.666 | -0.0134 | Yes |
| 97 | Eif4e | 11479 | -0.700 | 0.0113 | Yes |