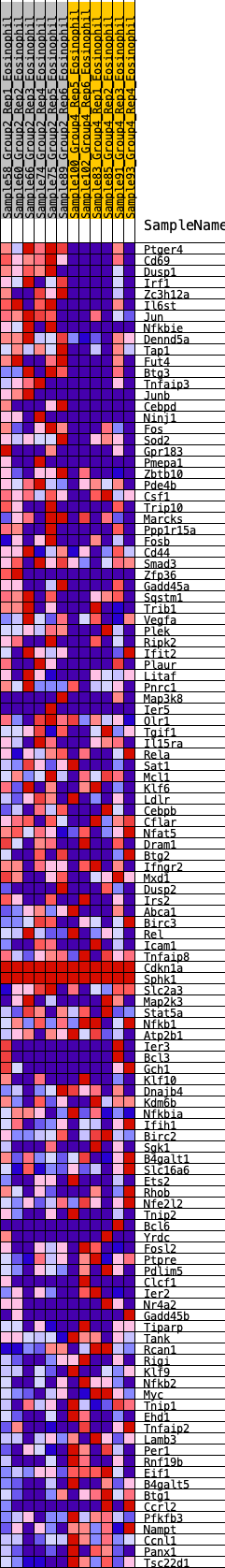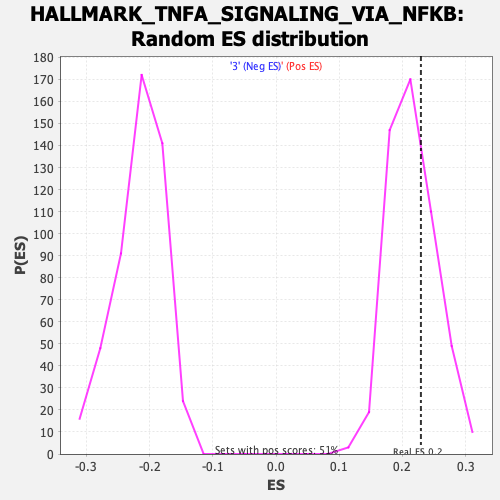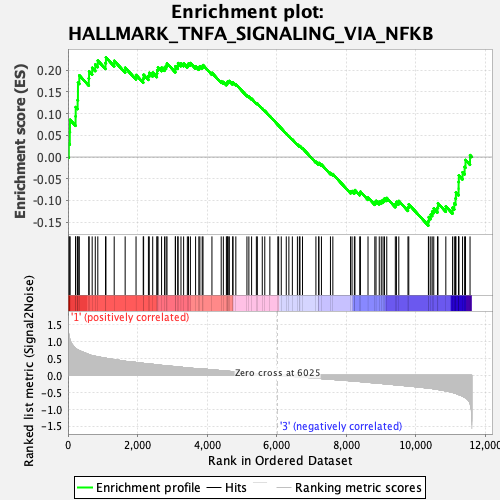
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group4.Eosinophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_TNFA_SIGNALING_VIA_NFKB |
| Enrichment Score (ES) | 0.22939745 |
| Normalized Enrichment Score (NES) | 1.0662698 |
| Nominal p-value | 0.32874015 |
| FDR q-value | 0.58733904 |
| FWER p-Value | 0.981 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ptger4 | 24 | 1.208 | 0.0308 | Yes |
| 2 | Cd69 | 52 | 1.072 | 0.0577 | Yes |
| 3 | Dusp1 | 57 | 1.045 | 0.0859 | Yes |
| 4 | Irf1 | 218 | 0.798 | 0.0937 | Yes |
| 5 | Zc3h12a | 222 | 0.796 | 0.1151 | Yes |
| 6 | Il6st | 277 | 0.756 | 0.1311 | Yes |
| 7 | Jun | 286 | 0.753 | 0.1509 | Yes |
| 8 | Nfkbie | 287 | 0.751 | 0.1714 | Yes |
| 9 | Dennd5a | 325 | 0.731 | 0.1881 | Yes |
| 10 | Tap1 | 598 | 0.618 | 0.1813 | Yes |
| 11 | Fut4 | 606 | 0.614 | 0.1974 | Yes |
| 12 | Btg3 | 695 | 0.580 | 0.2055 | Yes |
| 13 | Tnfaip3 | 784 | 0.563 | 0.2132 | Yes |
| 14 | Junb | 857 | 0.550 | 0.2220 | Yes |
| 15 | Cebpd | 1078 | 0.508 | 0.2167 | Yes |
| 16 | Ninj1 | 1091 | 0.505 | 0.2294 | Yes |
| 17 | Fos | 1328 | 0.467 | 0.2216 | No |
| 18 | Sod2 | 1643 | 0.419 | 0.2057 | No |
| 19 | Gpr183 | 1956 | 0.382 | 0.1889 | No |
| 20 | Pmepa1 | 2163 | 0.355 | 0.1807 | No |
| 21 | Zbtb10 | 2174 | 0.354 | 0.1895 | No |
| 22 | Pde4b | 2312 | 0.339 | 0.1868 | No |
| 23 | Csf1 | 2336 | 0.337 | 0.1940 | No |
| 24 | Trip10 | 2435 | 0.326 | 0.1944 | No |
| 25 | Marcks | 2555 | 0.312 | 0.1925 | No |
| 26 | Ppp1r15a | 2563 | 0.310 | 0.2003 | No |
| 27 | Fosb | 2587 | 0.308 | 0.2067 | No |
| 28 | Cd44 | 2690 | 0.295 | 0.2059 | No |
| 29 | Smad3 | 2776 | 0.287 | 0.2063 | No |
| 30 | Zfp36 | 2819 | 0.283 | 0.2104 | No |
| 31 | Gadd45a | 2846 | 0.282 | 0.2158 | No |
| 32 | Sqstm1 | 3086 | 0.259 | 0.2021 | No |
| 33 | Trib1 | 3088 | 0.259 | 0.2090 | No |
| 34 | Vegfa | 3156 | 0.251 | 0.2101 | No |
| 35 | Plek | 3164 | 0.251 | 0.2163 | No |
| 36 | Ripk2 | 3248 | 0.241 | 0.2156 | No |
| 37 | Ifit2 | 3324 | 0.232 | 0.2154 | No |
| 38 | Plaur | 3431 | 0.220 | 0.2122 | No |
| 39 | Litaf | 3464 | 0.216 | 0.2153 | No |
| 40 | Pnrc1 | 3519 | 0.213 | 0.2164 | No |
| 41 | Map3k8 | 3668 | 0.199 | 0.2090 | No |
| 42 | Ier5 | 3762 | 0.196 | 0.2062 | No |
| 43 | Olr1 | 3791 | 0.194 | 0.2091 | No |
| 44 | Tgif1 | 3858 | 0.190 | 0.2085 | No |
| 45 | Il15ra | 3881 | 0.187 | 0.2117 | No |
| 46 | Rela | 4138 | 0.164 | 0.1939 | No |
| 47 | Sat1 | 4404 | 0.138 | 0.1746 | No |
| 48 | Mcl1 | 4466 | 0.131 | 0.1728 | No |
| 49 | Klf6 | 4553 | 0.127 | 0.1688 | No |
| 50 | Ldlr | 4575 | 0.125 | 0.1704 | No |
| 51 | Cebpb | 4582 | 0.124 | 0.1733 | No |
| 52 | Cflar | 4606 | 0.122 | 0.1746 | No |
| 53 | Nfat5 | 4638 | 0.119 | 0.1751 | No |
| 54 | Dram1 | 4734 | 0.109 | 0.1698 | No |
| 55 | Btg2 | 4744 | 0.107 | 0.1720 | No |
| 56 | Ifngr2 | 4825 | 0.100 | 0.1678 | No |
| 57 | Mxd1 | 5144 | 0.075 | 0.1421 | No |
| 58 | Dusp2 | 5200 | 0.070 | 0.1392 | No |
| 59 | Irs2 | 5278 | 0.066 | 0.1343 | No |
| 60 | Abca1 | 5415 | 0.054 | 0.1240 | No |
| 61 | Birc3 | 5443 | 0.051 | 0.1230 | No |
| 62 | Rel | 5585 | 0.037 | 0.1117 | No |
| 63 | Icam1 | 5657 | 0.031 | 0.1064 | No |
| 64 | Tnfaip8 | 5803 | 0.019 | 0.0943 | No |
| 65 | Cdkn1a | 6033 | 0.000 | 0.0744 | No |
| 66 | Sphk1 | 6056 | 0.000 | 0.0724 | No |
| 67 | Slc2a3 | 6131 | -0.003 | 0.0661 | No |
| 68 | Map2k3 | 6275 | -0.015 | 0.0541 | No |
| 69 | Stat5a | 6350 | -0.021 | 0.0482 | No |
| 70 | Nfkb1 | 6450 | -0.029 | 0.0404 | No |
| 71 | Atp2b1 | 6595 | -0.040 | 0.0289 | No |
| 72 | Ier3 | 6651 | -0.046 | 0.0254 | No |
| 73 | Bcl3 | 6669 | -0.046 | 0.0252 | No |
| 74 | Gch1 | 6742 | -0.050 | 0.0203 | No |
| 75 | Klf10 | 7127 | -0.080 | -0.0109 | No |
| 76 | Dnajb4 | 7206 | -0.087 | -0.0154 | No |
| 77 | Kdm6b | 7214 | -0.087 | -0.0136 | No |
| 78 | Nfkbia | 7286 | -0.093 | -0.0172 | No |
| 79 | Ifih1 | 7545 | -0.113 | -0.0366 | No |
| 80 | Birc2 | 7614 | -0.119 | -0.0393 | No |
| 81 | Sgk1 | 8123 | -0.159 | -0.0791 | No |
| 82 | B4galt1 | 8166 | -0.163 | -0.0783 | No |
| 83 | Slc16a6 | 8233 | -0.169 | -0.0795 | No |
| 84 | Ets2 | 8249 | -0.170 | -0.0761 | No |
| 85 | Rhob | 8387 | -0.181 | -0.0831 | No |
| 86 | Nfe2l2 | 8407 | -0.183 | -0.0798 | No |
| 87 | Tnip2 | 8625 | -0.204 | -0.0931 | No |
| 88 | Bcl6 | 8821 | -0.220 | -0.1041 | No |
| 89 | Yrdc | 8855 | -0.222 | -0.1009 | No |
| 90 | Fosl2 | 8948 | -0.232 | -0.1026 | No |
| 91 | Ptpre | 9006 | -0.238 | -0.1010 | No |
| 92 | Pdlim5 | 9057 | -0.243 | -0.0988 | No |
| 93 | Clcf1 | 9098 | -0.247 | -0.0955 | No |
| 94 | Ier2 | 9164 | -0.253 | -0.0943 | No |
| 95 | Nr4a2 | 9410 | -0.276 | -0.1081 | No |
| 96 | Gadd45b | 9442 | -0.279 | -0.1032 | No |
| 97 | Tiparp | 9510 | -0.286 | -0.1012 | No |
| 98 | Tank | 9774 | -0.313 | -0.1156 | No |
| 99 | Rcan1 | 9797 | -0.315 | -0.1089 | No |
| 100 | Rigi | 10361 | -0.376 | -0.1476 | No |
| 101 | Klf9 | 10373 | -0.378 | -0.1383 | No |
| 102 | Nfkb2 | 10429 | -0.384 | -0.1326 | No |
| 103 | Myc | 10470 | -0.388 | -0.1255 | No |
| 104 | Tnip1 | 10515 | -0.396 | -0.1185 | No |
| 105 | Ehd1 | 10622 | -0.411 | -0.1166 | No |
| 106 | Tnfaip2 | 10636 | -0.413 | -0.1064 | No |
| 107 | Lamb3 | 10862 | -0.458 | -0.1135 | No |
| 108 | Per1 | 11056 | -0.502 | -0.1166 | No |
| 109 | Rnf19b | 11106 | -0.517 | -0.1068 | No |
| 110 | Eif1 | 11141 | -0.529 | -0.0953 | No |
| 111 | B4galt5 | 11152 | -0.532 | -0.0817 | No |
| 112 | Btg1 | 11229 | -0.565 | -0.0729 | No |
| 113 | Ccrl2 | 11230 | -0.566 | -0.0575 | No |
| 114 | Pfkfb3 | 11236 | -0.568 | -0.0424 | No |
| 115 | Nampt | 11343 | -0.608 | -0.0350 | No |
| 116 | Ccnl1 | 11403 | -0.643 | -0.0226 | No |
| 117 | Panx1 | 11428 | -0.661 | -0.0067 | No |
| 118 | Tsc22d1 | 11558 | -0.820 | 0.0044 | No |