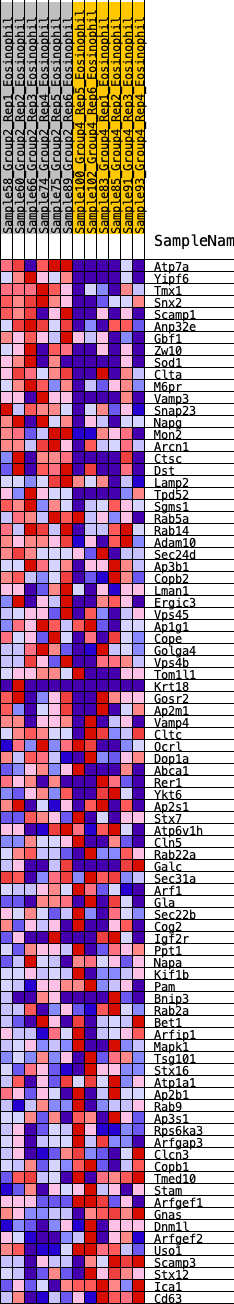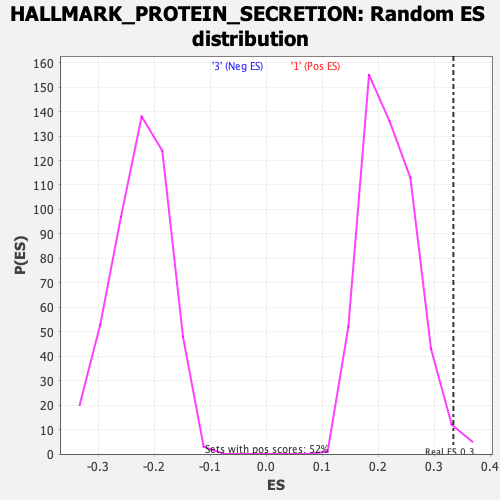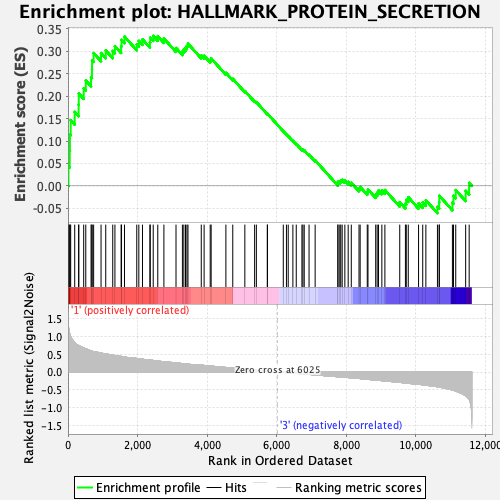
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group4.Eosinophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.33457112 |
| Normalized Enrichment Score (NES) | 1.5161703 |
| Nominal p-value | 0.023210831 |
| FDR q-value | 0.1465679 |
| FWER p-Value | 0.277 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Atp7a | 15 | 1.298 | 0.0436 | Yes |
| 2 | Yipf6 | 49 | 1.082 | 0.0782 | Yes |
| 3 | Tmx1 | 55 | 1.053 | 0.1142 | Yes |
| 4 | Snx2 | 77 | 0.990 | 0.1467 | Yes |
| 5 | Scamp1 | 193 | 0.818 | 0.1650 | Yes |
| 6 | Anp32e | 306 | 0.740 | 0.1809 | Yes |
| 7 | Gbf1 | 310 | 0.739 | 0.2062 | Yes |
| 8 | Zw10 | 451 | 0.676 | 0.2175 | Yes |
| 9 | Sod1 | 511 | 0.652 | 0.2349 | Yes |
| 10 | Clta | 663 | 0.591 | 0.2423 | Yes |
| 11 | M6pr | 689 | 0.582 | 0.2603 | Yes |
| 12 | Vamp3 | 692 | 0.581 | 0.2802 | Yes |
| 13 | Snap23 | 737 | 0.572 | 0.2962 | Yes |
| 14 | Napg | 950 | 0.531 | 0.2962 | Yes |
| 15 | Mon2 | 1085 | 0.506 | 0.3021 | Yes |
| 16 | Arcn1 | 1286 | 0.473 | 0.3011 | Yes |
| 17 | Ctsc | 1349 | 0.463 | 0.3117 | Yes |
| 18 | Dst | 1527 | 0.437 | 0.3115 | Yes |
| 19 | Lamp2 | 1539 | 0.436 | 0.3256 | Yes |
| 20 | Tpd52 | 1624 | 0.422 | 0.3330 | Yes |
| 21 | Sgms1 | 1977 | 0.379 | 0.3155 | Yes |
| 22 | Rab5a | 2036 | 0.373 | 0.3234 | Yes |
| 23 | Rab14 | 2141 | 0.358 | 0.3268 | Yes |
| 24 | Adam10 | 2355 | 0.335 | 0.3199 | Yes |
| 25 | Sec24d | 2364 | 0.333 | 0.3307 | Yes |
| 26 | Ap3b1 | 2450 | 0.324 | 0.3346 | Yes |
| 27 | Copb2 | 2581 | 0.308 | 0.3340 | No |
| 28 | Lman1 | 2757 | 0.290 | 0.3288 | No |
| 29 | Ergic3 | 3105 | 0.257 | 0.3076 | No |
| 30 | Vps45 | 3289 | 0.237 | 0.2999 | No |
| 31 | Ap1g1 | 3330 | 0.231 | 0.3044 | No |
| 32 | Cope | 3378 | 0.226 | 0.3082 | No |
| 33 | Golga4 | 3419 | 0.221 | 0.3123 | No |
| 34 | Vps4b | 3449 | 0.218 | 0.3174 | No |
| 35 | Tom1l1 | 3831 | 0.192 | 0.2909 | No |
| 36 | Krt18 | 3913 | 0.184 | 0.2903 | No |
| 37 | Gosr2 | 4088 | 0.168 | 0.2810 | No |
| 38 | Ap2m1 | 4115 | 0.165 | 0.2844 | No |
| 39 | Vamp4 | 4539 | 0.129 | 0.2522 | No |
| 40 | Cltc | 4737 | 0.108 | 0.2388 | No |
| 41 | Ocrl | 5084 | 0.080 | 0.2116 | No |
| 42 | Dop1a | 5365 | 0.058 | 0.1893 | No |
| 43 | Abca1 | 5415 | 0.054 | 0.1869 | No |
| 44 | Rer1 | 5730 | 0.024 | 0.1605 | No |
| 45 | Ykt6 | 5734 | 0.024 | 0.1610 | No |
| 46 | Ap2s1 | 6189 | -0.008 | 0.1219 | No |
| 47 | Stx7 | 6280 | -0.015 | 0.1146 | No |
| 48 | Atp6v1h | 6331 | -0.019 | 0.1110 | No |
| 49 | Cln5 | 6465 | -0.031 | 0.1005 | No |
| 50 | Rab22a | 6562 | -0.038 | 0.0935 | No |
| 51 | Galc | 6725 | -0.049 | 0.0811 | No |
| 52 | Sec31a | 6754 | -0.051 | 0.0805 | No |
| 53 | Arf1 | 6791 | -0.054 | 0.0792 | No |
| 54 | Gla | 6930 | -0.065 | 0.0695 | No |
| 55 | Sec22b | 7105 | -0.077 | 0.0570 | No |
| 56 | Cog2 | 7758 | -0.131 | 0.0050 | No |
| 57 | Igf2r | 7761 | -0.131 | 0.0093 | No |
| 58 | Ppt1 | 7814 | -0.135 | 0.0095 | No |
| 59 | Napa | 7842 | -0.138 | 0.0119 | No |
| 60 | Kif1b | 7882 | -0.141 | 0.0134 | No |
| 61 | Pam | 7959 | -0.149 | 0.0120 | No |
| 62 | Bnip3 | 8055 | -0.153 | 0.0090 | No |
| 63 | Rab2a | 8144 | -0.161 | 0.0070 | No |
| 64 | Bet1 | 8366 | -0.179 | -0.0060 | No |
| 65 | Arfip1 | 8399 | -0.182 | -0.0025 | No |
| 66 | Mapk1 | 8603 | -0.202 | -0.0131 | No |
| 67 | Tsg101 | 8624 | -0.204 | -0.0078 | No |
| 68 | Stx16 | 8846 | -0.221 | -0.0193 | No |
| 69 | Atp1a1 | 8888 | -0.226 | -0.0151 | No |
| 70 | Ap2b1 | 8927 | -0.230 | -0.0104 | No |
| 71 | Rab9 | 9023 | -0.240 | -0.0104 | No |
| 72 | Ap3s1 | 9110 | -0.248 | -0.0092 | No |
| 73 | Rps6ka3 | 9537 | -0.288 | -0.0362 | No |
| 74 | Arfgap3 | 9700 | -0.305 | -0.0398 | No |
| 75 | Clcn3 | 9725 | -0.308 | -0.0312 | No |
| 76 | Copb1 | 9782 | -0.313 | -0.0252 | No |
| 77 | Tmed10 | 10078 | -0.341 | -0.0390 | No |
| 78 | Stam | 10198 | -0.354 | -0.0371 | No |
| 79 | Arfgef1 | 10288 | -0.368 | -0.0321 | No |
| 80 | Gnas | 10621 | -0.411 | -0.0467 | No |
| 81 | Dnm1l | 10670 | -0.421 | -0.0363 | No |
| 82 | Arfgef2 | 10675 | -0.422 | -0.0220 | No |
| 83 | Uso1 | 11051 | -0.501 | -0.0373 | No |
| 84 | Scamp3 | 11082 | -0.511 | -0.0222 | No |
| 85 | Stx12 | 11147 | -0.530 | -0.0094 | No |
| 86 | Ica1 | 11432 | -0.663 | -0.0111 | No |
| 87 | Cd63 | 11533 | -0.762 | 0.0066 | No |