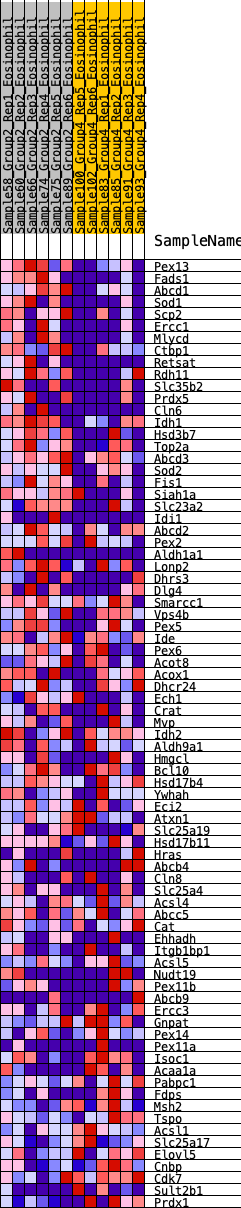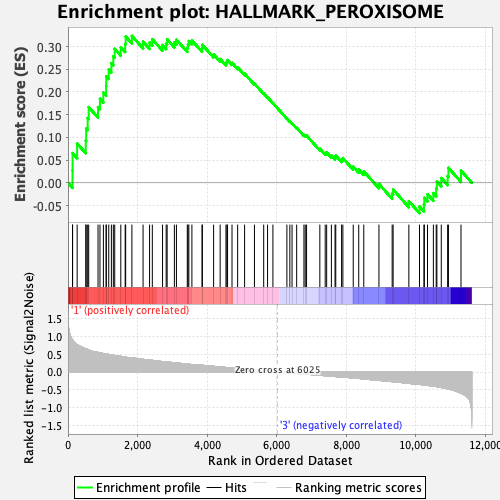
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Eosinophil.Eosinophil_Pheno.cls #Group2_versus_Group4.Eosinophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Eosinophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.3235511 |
| Normalized Enrichment Score (NES) | 1.3017577 |
| Nominal p-value | 0.10224949 |
| FDR q-value | 0.31226763 |
| FWER p-Value | 0.719 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pex13 | 130 | 0.901 | 0.0271 | Yes |
| 2 | Fads1 | 132 | 0.901 | 0.0654 | Yes |
| 3 | Abcd1 | 263 | 0.760 | 0.0865 | Yes |
| 4 | Sod1 | 511 | 0.652 | 0.0929 | Yes |
| 5 | Scp2 | 523 | 0.648 | 0.1195 | Yes |
| 6 | Ercc1 | 566 | 0.629 | 0.1427 | Yes |
| 7 | Mlycd | 595 | 0.619 | 0.1666 | Yes |
| 8 | Ctbp1 | 863 | 0.549 | 0.1669 | Yes |
| 9 | Retsat | 919 | 0.537 | 0.1850 | Yes |
| 10 | Rdh11 | 1017 | 0.518 | 0.1986 | Yes |
| 11 | Slc35b2 | 1096 | 0.504 | 0.2134 | Yes |
| 12 | Prdx5 | 1100 | 0.504 | 0.2346 | Yes |
| 13 | Cln6 | 1171 | 0.493 | 0.2495 | Yes |
| 14 | Idh1 | 1247 | 0.479 | 0.2634 | Yes |
| 15 | Hsd3b7 | 1303 | 0.470 | 0.2786 | Yes |
| 16 | Top2a | 1343 | 0.464 | 0.2950 | Yes |
| 17 | Abcd3 | 1518 | 0.439 | 0.2986 | Yes |
| 18 | Sod2 | 1643 | 0.419 | 0.3057 | Yes |
| 19 | Fis1 | 1658 | 0.414 | 0.3222 | Yes |
| 20 | Siah1a | 1837 | 0.395 | 0.3236 | Yes |
| 21 | Slc23a2 | 2158 | 0.355 | 0.3109 | No |
| 22 | Idi1 | 2345 | 0.336 | 0.3091 | No |
| 23 | Abcd2 | 2424 | 0.327 | 0.3163 | No |
| 24 | Pex2 | 2718 | 0.292 | 0.3033 | No |
| 25 | Aldh1a1 | 2822 | 0.283 | 0.3065 | No |
| 26 | Lonp2 | 2850 | 0.282 | 0.3161 | No |
| 27 | Dhrs3 | 3058 | 0.262 | 0.3093 | No |
| 28 | Dlg4 | 3117 | 0.255 | 0.3151 | No |
| 29 | Smarcc1 | 3429 | 0.220 | 0.2975 | No |
| 30 | Vps4b | 3449 | 0.218 | 0.3052 | No |
| 31 | Pex5 | 3474 | 0.215 | 0.3123 | No |
| 32 | Ide | 3562 | 0.209 | 0.3136 | No |
| 33 | Pex6 | 3853 | 0.190 | 0.2966 | No |
| 34 | Acot8 | 3863 | 0.189 | 0.3039 | No |
| 35 | Acox1 | 4186 | 0.159 | 0.2827 | No |
| 36 | Dhcr24 | 4374 | 0.141 | 0.2726 | No |
| 37 | Ech1 | 4541 | 0.128 | 0.2636 | No |
| 38 | Crat | 4568 | 0.125 | 0.2667 | No |
| 39 | Mvp | 4587 | 0.124 | 0.2704 | No |
| 40 | Idh2 | 4715 | 0.111 | 0.2641 | No |
| 41 | Aldh9a1 | 4877 | 0.096 | 0.2543 | No |
| 42 | Hmgcl | 5077 | 0.081 | 0.2405 | No |
| 43 | Bcl10 | 5360 | 0.059 | 0.2185 | No |
| 44 | Hsd17b4 | 5626 | 0.033 | 0.1969 | No |
| 45 | Ywhah | 5737 | 0.024 | 0.1884 | No |
| 46 | Eci2 | 5891 | 0.011 | 0.1756 | No |
| 47 | Atxn1 | 6293 | -0.016 | 0.1415 | No |
| 48 | Slc25a19 | 6378 | -0.024 | 0.1352 | No |
| 49 | Hsd17b11 | 6441 | -0.029 | 0.1311 | No |
| 50 | Hras | 6574 | -0.039 | 0.1213 | No |
| 51 | Abcb4 | 6783 | -0.053 | 0.1055 | No |
| 52 | Cln8 | 6820 | -0.057 | 0.1048 | No |
| 53 | Slc25a4 | 6858 | -0.060 | 0.1042 | No |
| 54 | Acsl4 | 7240 | -0.090 | 0.0749 | No |
| 55 | Abcc5 | 7394 | -0.101 | 0.0660 | No |
| 56 | Cat | 7436 | -0.104 | 0.0669 | No |
| 57 | Ehhadh | 7572 | -0.116 | 0.0601 | No |
| 58 | Itgb1bp1 | 7677 | -0.124 | 0.0563 | No |
| 59 | Acsl5 | 7698 | -0.126 | 0.0600 | No |
| 60 | Nudt19 | 7865 | -0.139 | 0.0515 | No |
| 61 | Pex11b | 7902 | -0.143 | 0.0545 | No |
| 62 | Abcb9 | 8200 | -0.166 | 0.0358 | No |
| 63 | Ercc3 | 8358 | -0.179 | 0.0298 | No |
| 64 | Gnpat | 8503 | -0.192 | 0.0255 | No |
| 65 | Pex14 | 8939 | -0.231 | -0.0024 | No |
| 66 | Pex11a | 9319 | -0.267 | -0.0239 | No |
| 67 | Isoc1 | 9347 | -0.271 | -0.0147 | No |
| 68 | Acaa1a | 9799 | -0.315 | -0.0404 | No |
| 69 | Pabpc1 | 10105 | -0.345 | -0.0521 | No |
| 70 | Fdps | 10233 | -0.360 | -0.0478 | No |
| 71 | Msh2 | 10244 | -0.363 | -0.0332 | No |
| 72 | Tspo | 10335 | -0.373 | -0.0251 | No |
| 73 | Acsl1 | 10504 | -0.394 | -0.0229 | No |
| 74 | Slc25a17 | 10585 | -0.405 | -0.0126 | No |
| 75 | Elovl5 | 10605 | -0.408 | 0.0031 | No |
| 76 | Cnbp | 10731 | -0.431 | 0.0106 | No |
| 77 | Cdk7 | 10914 | -0.468 | 0.0148 | No |
| 78 | Sult2b1 | 10937 | -0.473 | 0.0330 | No |
| 79 | Prdx1 | 11298 | -0.590 | 0.0270 | No |